Please be patient as the page loads
|
DACT1_MOUSE
|
||||||
| SwissProt Accessions | Q8R4A3, Q80VG9, Q8BP49, Q9JK89 | Gene names | Dact1, Thyex3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (MDpr1) (Frodo) (Thymus-expressed novel gene 3 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DACT1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.898184 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
DACT1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8R4A3, Q80VG9, Q8BP49, Q9JK89 | Gene names | Dact1, Thyex3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (MDpr1) (Frodo) (Thymus-expressed novel gene 3 protein). | |||||
|
CAP2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 3) | NC score | 0.112613 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |
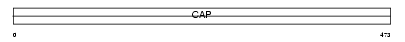
|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 4) | NC score | 0.129548 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.071377 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 6) | NC score | 0.081409 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.051508 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.021397 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.065775 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
FBRL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.060584 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35550, Q99L58 | Gene names | Fbl | |||
|
Domain Architecture |
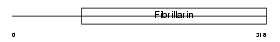
|
|||||
| Description | rRNA 2'-O-methyltransferase fibrillarin (EC 2.1.1.-) (Nucleolar protein 1). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.014732 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.005504 (rank : 96) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.019599 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.041082 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.036783 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.030245 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.058404 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.026344 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
NECD_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.022042 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.075183 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.017378 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
ZF106_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.029328 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.049259 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.018156 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.019767 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.040781 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.023950 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.023364 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.071110 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.039195 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.014334 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.019926 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CS016_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.035499 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.010022 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
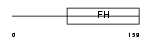
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.021906 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.049059 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NUMBL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.021742 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
RRS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.031932 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15050, Q9BUX8 | Gene names | RRS1, KIAA0112, RRR | |||
|
Domain Architecture |
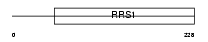
|
|||||
| Description | Ribosome biogenesis regulatory protein homolog. | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.046283 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
CAP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.059910 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P40124 | Gene names | Cap1, Cap | |||
|
Domain Architecture |
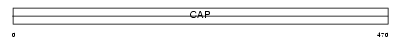
|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.025563 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
FA53C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.026046 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYF3 | Gene names | FAM53C, C5orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM53C. | |||||
|
GP179_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.026031 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.023098 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYPN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.009287 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.054388 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PAK6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.002077 (rank : 98) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQU5 | Gene names | PAK6, PAK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 6 (EC 2.7.11.1) (p21-activated kinase 6) (PAK-6) (PAK-5). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.038039 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.024954 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SETB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.018313 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.027204 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.008193 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.044695 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.018570 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.029464 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.023933 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
NAL10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.020683 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
NCOA5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.022900 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.021266 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
OTU7A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.017574 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.023520 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
PRDM8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.015443 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQV8, Q6IQ36 | Gene names | PRDM8, PFM5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 8 (PR domain-containing protein 8). | |||||
|
REST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.025276 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
TBCD5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.018883 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XQ2 | Gene names | Tbc1d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 5. | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.081529 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.040423 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CAP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.060614 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYT6, Q80YU7, Q9D6L0 | Gene names | Cap2 | |||
|
Domain Architecture |
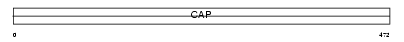
|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CREG2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.028497 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGC9, Q8BL64, Q8BX89, Q8K049 | Gene names | Creg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CREG2 precursor. | |||||
|
DPOLL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.017864 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXE2, Q9CTJ1 | Gene names | Poll, Polk | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase lambda (EC 2.7.7.7) (EC 4.2.99.-) (Pol Lambda) (DNA polymerase kappa). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.019102 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.019285 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.009225 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.021980 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.021857 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
RHG12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.015954 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
SUZ12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.027497 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15022, Q96BD9 | Gene names | SUZ12, CHET9, JJAZ1, KIAA0160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SUZ12 (Suppressor of zeste 12 protein homolog) (Joined to JAZF1 protein) (Chromatin precipitated E2F target 9 protein) (ChET 9 protein). | |||||
|
SUZ12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.028419 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U70, Q80Y10, Q99L07 | Gene names | Suz12, D11Ertd530e, Kiaa0160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein Suz12 (Suppressor of zeste 12 protein homolog). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.027894 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.042749 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.017981 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.006050 (rank : 95) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.034894 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.023206 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.019599 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CN102_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.016450 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H7Z3, Q9NWH6 | Gene names | C14orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf102. | |||||
|
INVS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.013401 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.042285 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MUCEN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.020704 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.022572 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NKX32_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.004086 (rank : 97) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78367 | Gene names | BAPX1, NKX3B | |||
|
Domain Architecture |
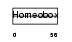
|
|||||
| Description | Homeobox protein Nkx-3.2 (Bagpipe homeobox protein homolog 1). | |||||
|
REST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.023553 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SNUT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.017200 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z315, Q8K155, Q9R1I9, Q9R270 | Gene names | Sart1, Haf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (Squamous cell carcinoma antigen recognized by T-cells 1) (SART-1) (mSART-1) (Hypoxia- associated factor). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.012548 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.077672 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.068267 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.057302 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.053074 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.061879 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
DACT1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8R4A3, Q80VG9, Q8BP49, Q9JK89 | Gene names | Dact1, Thyex3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (MDpr1) (Frodo) (Thymus-expressed novel gene 3 protein). | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.898184 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.129548 (rank : 3) | θ value | 0.0113563 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
CAP2_HUMAN
|
||||||
| NC score | 0.112613 (rank : 4) | θ value | 0.00298849 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.081529 (rank : 5) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.081409 (rank : 6) | θ value | 0.0148317 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.077672 (rank : 7) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.075183 (rank : 8) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.071377 (rank : 9) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.071110 (rank : 10) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.068267 (rank : 11) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.065775 (rank : 12) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.061879 (rank : 13) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CAP2_MOUSE
|
||||||
| NC score | 0.060614 (rank : 14) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYT6, Q80YU7, Q9D6L0 | Gene names | Cap2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
FBRL_MOUSE
|
||||||
| NC score | 0.060584 (rank : 15) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35550, Q99L58 | Gene names | Fbl | |||
|
Domain Architecture |

|
|||||
| Description | rRNA 2'-O-methyltransferase fibrillarin (EC 2.1.1.-) (Nucleolar protein 1). | |||||
|
CAP1_MOUSE
|
||||||
| NC score | 0.059910 (rank : 16) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P40124 | Gene names | Cap1, Cap | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.058404 (rank : 17) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.057302 (rank : 18) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.054388 (rank : 19) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.053074 (rank : 20) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.051508 (rank : 21) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.049259 (rank : 22) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.049059 (rank : 23) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.046283 (rank : 24) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.044695 (rank : 25) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.042749 (rank : 26) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.042285 (rank : 27) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.041082 (rank : 28) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.040781 (rank : 29) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.040423 (rank : 30) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.039195 (rank : 31) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.038039 (rank : 32) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.036783 (rank : 33) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.035499 (rank : 34) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.034894 (rank : 35) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
RRS1_HUMAN
|
||||||
| NC score | 0.031932 (rank : 36) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15050, Q9BUX8 | Gene names | RRS1, KIAA0112, RRR | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis regulatory protein homolog. | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.030245 (rank : 37) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.029464 (rank : 38) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
ZF106_HUMAN
|
||||||
| NC score | 0.029328 (rank : 39) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
CREG2_MOUSE
|
||||||
| NC score | 0.028497 (rank : 40) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGC9, Q8BL64, Q8BX89, Q8K049 | Gene names | Creg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CREG2 precursor. | |||||
|
SUZ12_MOUSE
|
||||||
| NC score | 0.028419 (rank : 41) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U70, Q80Y10, Q99L07 | Gene names | Suz12, D11Ertd530e, Kiaa0160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein Suz12 (Suppressor of zeste 12 protein homolog). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.027894 (rank : 42) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SUZ12_HUMAN
|
||||||
| NC score | 0.027497 (rank : 43) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15022, Q96BD9 | Gene names | SUZ12, CHET9, JJAZ1, KIAA0160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SUZ12 (Suppressor of zeste 12 protein homolog) (Joined to JAZF1 protein) (Chromatin precipitated E2F target 9 protein) (ChET 9 protein). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.027204 (rank : 44) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.026344 (rank : 45) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
FA53C_HUMAN
|
||||||
| NC score | 0.026046 (rank : 46) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYF3 | Gene names | FAM53C, C5orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM53C. | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.026031 (rank : 47) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.025563 (rank : 48) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.025276 (rank : 49) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.024954 (rank : 50) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.023950 (rank : 51) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.023933 (rank : 52) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.023553 (rank : 53) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.023520 (rank : 54) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.023364 (rank : 55) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.023206 (rank : 56) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.023098 (rank : 57) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NCOA5_HUMAN
|
||||||
| NC score | 0.022900 (rank : 58) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.022572 (rank : 59) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NECD_MOUSE
|
||||||
| NC score | 0.022042 (rank : 60) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25233, Q542W7, Q61951 | Gene names | Ndn | |||
|
Domain Architecture |

|
|||||
| Description | Necdin. | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.021980 (rank : 61) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.021906 (rank : 62) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.021857 (rank : 63) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
NUMBL_MOUSE
|
||||||
| NC score | 0.021742 (rank : 64) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.021397 (rank : 65) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.021266 (rank : 66) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
MUCEN_MOUSE
|
||||||
| NC score | 0.020704 (rank : 67) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
NAL10_MOUSE
|
||||||
| NC score | 0.020683 (rank : 68) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.019926 (rank : 69) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.019767 (rank : 70) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.019599 (rank : 71) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.019599 (rank : 72) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.019285 (rank : 73) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.019102 (rank : 74) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
TBCD5_MOUSE
|
||||||
| NC score | 0.018883 (rank : 75) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XQ2 | Gene names | Tbc1d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 5. | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.018570 (rank : 76) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
SETB1_MOUSE
|
||||||
| NC score | 0.018313 (rank : 77) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.018156 (rank : 78) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.017981 (rank : 79) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
DPOLL_MOUSE
|
||||||
| NC score | 0.017864 (rank : 80) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXE2, Q9CTJ1 | Gene names | Poll, Polk | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase lambda (EC 2.7.7.7) (EC 4.2.99.-) (Pol Lambda) (DNA polymerase kappa). | |||||
|
OTU7A_HUMAN
|
||||||
| NC score | 0.017574 (rank : 81) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.017378 (rank : 82) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
SNUT1_MOUSE
|
||||||
| NC score | 0.017200 (rank : 83) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z315, Q8K155, Q9R1I9, Q9R270 | Gene names | Sart1, Haf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (Squamous cell carcinoma antigen recognized by T-cells 1) (SART-1) (mSART-1) (Hypoxia- associated factor). | |||||
|
CN102_HUMAN
|
||||||
| NC score | 0.016450 (rank : 84) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H7Z3, Q9NWH6 | Gene names | C14orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf102. | |||||
|
RHG12_HUMAN
|
||||||
| NC score | 0.015954 (rank : 85) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWW6, Q86UB3, Q8IWW7, Q8N3L1, Q9NT76 | Gene names | ARHGAP12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
PRDM8_HUMAN
|
||||||
| NC score | 0.015443 (rank : 86) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQV8, Q6IQ36 | Gene names | PRDM8, PFM5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 8 (PR domain-containing protein 8). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.014732 (rank : 87) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.014334 (rank : 88) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.013401 (rank : 89) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.012548 (rank : 90) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.010022 (rank : 91) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
MYPN_HUMAN
|
||||||
| NC score | 0.009287 (rank : 92) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.009225 (rank : 93) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.008193 (rank : 94) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.006050 (rank : 95) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.005504 (rank : 96) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
NKX32_HUMAN
|
||||||
| NC score | 0.004086 (rank : 97) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78367 | Gene names | BAPX1, NKX3B | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.2 (Bagpipe homeobox protein homolog 1). | |||||
|
PAK6_HUMAN
|
||||||
| NC score | 0.002077 (rank : 98) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQU5 | Gene names | PAK6, PAK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 6 (EC 2.7.11.1) (p21-activated kinase 6) (PAK-6) (PAK-5). | |||||