Please be patient as the page loads
|
SMAD5_HUMAN
|
||||||
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMAD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994862 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995033 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998440 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD9_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.994002 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD9_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.989779 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD3_HUMAN
|
||||||
| θ value | 2.8663e-147 (rank : 7) | NC score | 0.986487 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| θ value | 2.8663e-147 (rank : 8) | NC score | 0.986487 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_HUMAN
|
||||||
| θ value | 9.56396e-135 (rank : 9) | NC score | 0.983116 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD2_MOUSE
|
||||||
| θ value | 9.56396e-135 (rank : 10) | NC score | 0.983119 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 2.4834e-66 (rank : 11) | NC score | 0.923893 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD4_HUMAN
|
||||||
| θ value | 7.2256e-66 (rank : 12) | NC score | 0.925775 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD6_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 13) | NC score | 0.767494 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD6_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 14) | NC score | 0.764605 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |
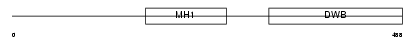
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD7_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 15) | NC score | 0.749850 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |
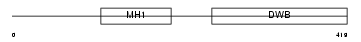
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
SMAD7_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 16) | NC score | 0.750645 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |
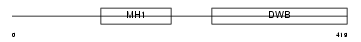
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.101271 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.048796 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.020113 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FOXJ1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.019210 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |
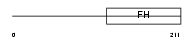
|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
EVX1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.016144 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49640 | Gene names | EVX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 1 (EVX-1). | |||||
|
EVX1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.016139 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23683 | Gene names | Evx1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 1 (EVX-1). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.010500 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
KBTB4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.022478 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NVX7, Q6IA85, Q9BUC3, Q9NV76 | Gene names | KBTBD4, BKLHD4 | |||
|
Domain Architecture |
|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
IRAK3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.006758 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.018591 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.022605 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
IRF4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.048313 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15306, Q99660 | Gene names | IRF4, MUM1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (Multiple myeloma oncogene 1). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.017104 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.013640 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
DAZ1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.041035 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
DAZ2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.041618 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
DAZ3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.041775 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
DAZ4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.041197 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
K0256_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.024672 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q93073, Q8N767 | Gene names | KIAA0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
KBTB4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.012840 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R179, Q9CQX1 | Gene names | Kbtbd4, Bklhd4 | |||
|
Domain Architecture |
|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.019460 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
K0256_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.023746 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6A098 | Gene names | Kiaa0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.011649 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.019325 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
P73L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.016467 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
P73L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.016561 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
BOP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.010924 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97452, Q3TK87, Q91X31 | Gene names | Bop1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BOP1 (Block of proliferation 1 protein). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | -0.000311 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
IRF7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.019647 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.030948 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TENS4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.015804 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.018701 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CDON_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.001952 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q32MD9, O88971 | Gene names | Cdon, Cdo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.012455 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.012816 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.007128 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.040862 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | -0.002582 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
HXD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.003280 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
LIPB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.002269 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.031853 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
SMAD5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| NC score | 0.998440 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD1_MOUSE
|
||||||
| NC score | 0.995033 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD1_HUMAN
|
||||||
| NC score | 0.994862 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD9_HUMAN
|
||||||
| NC score | 0.994002 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD9_MOUSE
|
||||||
| NC score | 0.989779 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD3_HUMAN
|
||||||
| NC score | 0.986487 (rank : 7) | θ value | 2.8663e-147 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| NC score | 0.986487 (rank : 8) | θ value | 2.8663e-147 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_MOUSE
|
||||||
| NC score | 0.983119 (rank : 9) | θ value | 9.56396e-135 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD2_HUMAN
|
||||||
| NC score | 0.983116 (rank : 10) | θ value | 9.56396e-135 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD4_HUMAN
|
||||||
| NC score | 0.925775 (rank : 11) | θ value | 7.2256e-66 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.923893 (rank : 12) | θ value | 2.4834e-66 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD6_MOUSE
|
||||||
| NC score | 0.767494 (rank : 13) | θ value | 8.65492e-19 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD6_HUMAN
|
||||||
| NC score | 0.764605 (rank : 14) | θ value | 2.5182e-18 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD7_MOUSE
|
||||||
| NC score | 0.750645 (rank : 15) | θ value | 4.44505e-15 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
SMAD7_HUMAN
|
||||||
| NC score | 0.749850 (rank : 16) | θ value | 1.52774e-15 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.101271 (rank : 17) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.048796 (rank : 18) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
IRF4_HUMAN
|
||||||
| NC score | 0.048313 (rank : 19) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15306, Q99660 | Gene names | IRF4, MUM1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (Multiple myeloma oncogene 1). | |||||
|
DAZ3_HUMAN
|
||||||
| NC score | 0.041775 (rank : 20) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
DAZ2_HUMAN
|
||||||
| NC score | 0.041618 (rank : 21) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
DAZ4_HUMAN
|
||||||
| NC score | 0.041197 (rank : 22) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
DAZ1_HUMAN
|
||||||
| NC score | 0.041035 (rank : 23) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.040862 (rank : 24) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.031853 (rank : 25) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.030948 (rank : 26) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
K0256_HUMAN
|
||||||
| NC score | 0.024672 (rank : 27) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q93073, Q8N767 | Gene names | KIAA0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
K0256_MOUSE
|
||||||
| NC score | 0.023746 (rank : 28) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6A098 | Gene names | Kiaa0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.022605 (rank : 29) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
KBTB4_HUMAN
|
||||||
| NC score | 0.022478 (rank : 30) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NVX7, Q6IA85, Q9BUC3, Q9NV76 | Gene names | KBTBD4, BKLHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.020113 (rank : 31) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
IRF7_HUMAN
|
||||||
| NC score | 0.019647 (rank : 32) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.019460 (rank : 33) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.019325 (rank : 34) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
FOXJ1_MOUSE
|
||||||
| NC score | 0.019210 (rank : 35) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.018701 (rank : 36) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.018591 (rank : 37) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.017104 (rank : 38) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
P73L_MOUSE
|
||||||
| NC score | 0.016561 (rank : 39) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73L_HUMAN
|
||||||
| NC score | 0.016467 (rank : 40) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
EVX1_HUMAN
|
||||||
| NC score | 0.016144 (rank : 41) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49640 | Gene names | EVX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 1 (EVX-1). | |||||
|
EVX1_MOUSE
|
||||||
| NC score | 0.016139 (rank : 42) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23683 | Gene names | Evx1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 1 (EVX-1). | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.015804 (rank : 43) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.013640 (rank : 44) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
KBTB4_MOUSE
|
||||||
| NC score | 0.012840 (rank : 45) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R179, Q9CQX1 | Gene names | Kbtbd4, Bklhd4 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.012816 (rank : 46) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.012455 (rank : 47) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.011649 (rank : 48) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
BOP1_MOUSE
|
||||||
| NC score | 0.010924 (rank : 49) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97452, Q3TK87, Q91X31 | Gene names | Bop1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BOP1 (Block of proliferation 1 protein). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.010500 (rank : 50) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.007128 (rank : 51) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
IRAK3_MOUSE
|
||||||
| NC score | 0.006758 (rank : 52) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 628 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4B2, Q8C7U8, Q8CE40, Q8K1S8 | Gene names | Irak3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 3 (EC 2.7.11.1) (IRAK-3) (IL- 1 receptor-associated kinase M) (IRAK-M). | |||||
|
HXD3_HUMAN
|
||||||
| NC score | 0.003280 (rank : 53) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31249, Q99955, Q9BSC5 | Gene names | HOXD3, HOX4A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4A). | |||||
|
LIPB2_HUMAN
|
||||||
| NC score | 0.002269 (rank : 54) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 599 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8ND30, O75337, Q8WW26 | Gene names | PPFIBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2). | |||||
|
CDON_MOUSE
|
||||||
| NC score | 0.001952 (rank : 55) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q32MD9, O88971 | Gene names | Cdon, Cdo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | -0.000311 (rank : 56) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | -0.002582 (rank : 57) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||