Please be patient as the page loads
|
K0256_MOUSE
|
||||||
| SwissProt Accessions | Q6A098 | Gene names | Kiaa0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K0256_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.973275 (rank : 2) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q93073, Q8N767 | Gene names | KIAA0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
K0256_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q6A098 | Gene names | Kiaa0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
SEBP2_HUMAN
|
||||||
| θ value | 3.82246e-75 (rank : 3) | NC score | 0.829823 (rank : 3) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
NF2L1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 4) | NC score | 0.062797 (rank : 6) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 5) | NC score | 0.055456 (rank : 12) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.066349 (rank : 5) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
NP1L3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.048088 (rank : 18) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.017511 (rank : 75) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.037739 (rank : 29) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.044269 (rank : 20) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.032685 (rank : 36) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.042146 (rank : 25) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.059557 (rank : 8) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.062183 (rank : 7) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.021068 (rank : 60) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CK035_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.047741 (rank : 19) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q0VET5, Q9CW10 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35 homolog. | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.012188 (rank : 87) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.055954 (rank : 11) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.020047 (rank : 63) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.069251 (rank : 4) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.053155 (rank : 15) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.043076 (rank : 24) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
SETB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.057797 (rank : 9) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.051278 (rank : 16) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.032123 (rank : 38) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TLE2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.018508 (rank : 69) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVB2 | Gene names | Tle2, Grg2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2. | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.043861 (rank : 23) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.039423 (rank : 27) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.032413 (rank : 37) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
NF2L1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.048619 (rank : 17) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.036104 (rank : 30) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SLC2B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.053279 (rank : 14) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
ARAF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.005140 (rank : 115) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.032102 (rank : 39) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BLNK_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.033751 (rank : 33) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
MCM3A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.030035 (rank : 44) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.057502 (rank : 10) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.017975 (rank : 70) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.017036 (rank : 77) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
SNRK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.004440 (rank : 117) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDU5, Q91WX6 | Gene names | Snrk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
BNC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.029123 (rank : 45) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
BNC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.028921 (rank : 46) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
INGR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.030308 (rank : 43) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15261 | Gene names | Ifngr1, Ifngr | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-gamma receptor alpha chain precursor (IFN-gamma-R1) (CD119 antigen). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.025013 (rank : 49) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
BAALC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.034091 (rank : 31) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHV1, Q8VBS8, Q9CYS9 | Gene names | Baalc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain and acute leukemia cytoplasmic protein. | |||||
|
GAK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.012471 (rank : 86) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
KS6C1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.009884 (rank : 98) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BLK9, Q3UDY3, Q6PDY4, Q8BMQ5, Q8BX49 | Gene names | Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase). | |||||
|
NFL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.008113 (rank : 106) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.005481 (rank : 114) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SMAD5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.023746 (rank : 54) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.023318 (rank : 55) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.054439 (rank : 13) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SUHW4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.008758 (rank : 102) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.017644 (rank : 72) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
BASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.033696 (rank : 34) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.028796 (rank : 47) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CABYR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.027282 (rank : 48) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
HIRA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.011725 (rank : 90) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61666, O08845, Q62365 | Gene names | Hira, Tuple1 | |||
|
Domain Architecture |
|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
RL7A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.041903 (rank : 26) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62424, P11518 | Gene names | RPL7A, SURF-3, SURF3 | |||
|
Domain Architecture |
|
|||||
| Description | 60S ribosomal protein L7a (Surfeit locus protein 3) (PLA-X polypeptide). | |||||
|
TLE2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.015145 (rank : 81) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.014857 (rank : 82) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
VP13B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.024552 (rank : 51) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7G8, Q709C6, Q709C7, Q7Z7G4, Q7Z7G5, Q7Z7G6, Q7Z7G7, Q8NB77, Q9NWV1, Q9Y4E7 | Gene names | VPS13B, CHS1, COH1, KIAA0532 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 13B (Cohen syndrome protein 1). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.034009 (rank : 32) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.012704 (rank : 85) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
BGLR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.017603 (rank : 73) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12265, Q61601, Q64473, Q64474 | Gene names | Gusb, Gus, Gus-s | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31). | |||||
|
KAISO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.005051 (rank : 116) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86T24, O00319, Q7Z361, Q8IVP6, Q8N3P0 | Gene names | ZBTB33, KAISO, ZNF348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.037770 (rank : 28) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.032097 (rank : 40) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PJA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.014268 (rank : 83) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.021621 (rank : 59) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.019812 (rank : 64) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.023801 (rank : 52) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.023801 (rank : 53) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SNRK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.003188 (rank : 119) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRH2, Q14706, Q68D15, Q6IQ46, Q9NXI7 | Gene names | SNRK, KIAA0096, SNFRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.021867 (rank : 58) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.019718 (rank : 65) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.022209 (rank : 57) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.032916 (rank : 35) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.011694 (rank : 91) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
FGD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.006969 (rank : 112) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
KCT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.020187 (rank : 62) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K201, Q5SVC5, Q922L7, Q9CVN1 | Gene names | Kct2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratinocytes-associated transmembrane protein 2 precursor. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.031610 (rank : 41) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.022341 (rank : 56) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.024898 (rank : 50) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.007501 (rank : 110) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.020670 (rank : 61) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PHF13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.017572 (rank : 74) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2W6 | Gene names | Phf13 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 13. | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.016051 (rank : 78) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RIMB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.019379 (rank : 66) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.015323 (rank : 80) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
XYLT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.011073 (rank : 94) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.019099 (rank : 68) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
AP180_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.019280 (rank : 67) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.009960 (rank : 97) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.011225 (rank : 93) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CS029_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.031598 (rank : 42) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.003691 (rank : 118) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CYB5B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.013756 (rank : 84) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQX2, Q9D1M6, Q9D8R3 | Gene names | Cyb5b, Cyb5m | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome b5 type B precursor (Cytochrome b5 outer mitochondrial membrane isoform). | |||||
|
DBND2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.017278 (rank : 76) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |
|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.005694 (rank : 113) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.009610 (rank : 100) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.011981 (rank : 89) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.010894 (rank : 95) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GTF2I_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.011277 (rank : 92) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.008247 (rank : 105) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
IRAK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.002227 (rank : 120) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |
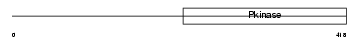
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.007321 (rank : 111) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
LPIN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.008675 (rank : 103) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |
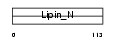
|
|||||
| Description | Lipin-3. | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.007805 (rank : 108) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NHPX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.044059 (rank : 21) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55769 | Gene names | NHP2L1 | |||
|
Domain Architecture |

|
|||||
| Description | NHP2-like protein 1 (High mobility group-like nuclear protein 2 homolog 1) (U4/U6.U5 tri-snRNP 15.5 kDa protein) (OTK27) (hSNU13). | |||||
|
NHPX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.044059 (rank : 22) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0T1, O08754, Q3UY64 | Gene names | Nhp2l1, Ssfa1 | |||
|
Domain Architecture |

|
|||||
| Description | NHP2-like protein 1 (High mobility group-like nuclear protein 2 homolog 1) (U4/U6.U5 tri-snRNP 15.5 kDa protein) (Sperm-specific antigen 1) (Fertilization antigen 1) (FA-1). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.012001 (rank : 88) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PI3R4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.008826 (rank : 101) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VD65, Q56A65, Q8C948, Q8C9D7, Q9CVC5 | Gene names | Pik3r4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 4 (EC 2.7.11.1) (PI3- kinase regulatory subunit 4). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.010841 (rank : 96) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.007628 (rank : 109) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RIMB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.017706 (rank : 71) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U40 | Gene names | Rimbp2, Kiaa0318, Rbp2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
RPGF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.009656 (rank : 99) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.008666 (rank : 104) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.015979 (rank : 79) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ZHX3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.007884 (rank : 107) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0Q2, Q80U14 | Gene names | Zhx3, Kiaa0395, Tix1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and homeodomain protein 3 (Triple homeobox protein 1). | |||||
|
ZSCA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | -0.001098 (rank : 121) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |
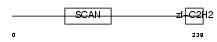
|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||
|
K0256_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q6A098 | Gene names | Kiaa0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
K0256_HUMAN
|
||||||
| NC score | 0.973275 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q93073, Q8N767 | Gene names | KIAA0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
SEBP2_HUMAN
|
||||||
| NC score | 0.829823 (rank : 3) | θ value | 3.82246e-75 (rank : 3) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.069251 (rank : 4) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.066349 (rank : 5) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
NF2L1_MOUSE
|
||||||
| NC score | 0.062797 (rank : 6) | θ value | 0.0148317 (rank : 4) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.062183 (rank : 7) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.059557 (rank : 8) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SETB2_HUMAN
|
||||||
| NC score | 0.057797 (rank : 9) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.057502 (rank : 10) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.055954 (rank : 11) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.055456 (rank : 12) | θ value | 0.0431538 (rank : 5) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.054439 (rank : 13) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SLC2B_HUMAN
|
||||||
| NC score | 0.053279 (rank : 14) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEV8, Q9Y4D6 | Gene names | EXPH5, KIAA0624, SLAC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Slp homolog lacking C2 domains b (Exophilin-5). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.053155 (rank : 15) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.051278 (rank : 16) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
NF2L1_HUMAN
|
||||||
| NC score | 0.048619 (rank : 17) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NP1L3_HUMAN
|
||||||
| NC score | 0.048088 (rank : 18) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
CK035_MOUSE
|
||||||
| NC score | 0.047741 (rank : 19) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q0VET5, Q9CW10 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35 homolog. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.044269 (rank : 20) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NHPX_HUMAN
|
||||||
| NC score | 0.044059 (rank : 21) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55769 | Gene names | NHP2L1 | |||
|
Domain Architecture |

|
|||||
| Description | NHP2-like protein 1 (High mobility group-like nuclear protein 2 homolog 1) (U4/U6.U5 tri-snRNP 15.5 kDa protein) (OTK27) (hSNU13). | |||||
|
NHPX_MOUSE
|
||||||
| NC score | 0.044059 (rank : 22) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0T1, O08754, Q3UY64 | Gene names | Nhp2l1, Ssfa1 | |||
|
Domain Architecture |

|
|||||
| Description | NHP2-like protein 1 (High mobility group-like nuclear protein 2 homolog 1) (U4/U6.U5 tri-snRNP 15.5 kDa protein) (Sperm-specific antigen 1) (Fertilization antigen 1) (FA-1). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.043861 (rank : 23) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.043076 (rank : 24) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.042146 (rank : 25) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
RL7A_HUMAN
|
||||||
| NC score | 0.041903 (rank : 26) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62424, P11518 | Gene names | RPL7A, SURF-3, SURF3 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L7a (Surfeit locus protein 3) (PLA-X polypeptide). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.039423 (rank : 27) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.037770 (rank : 28) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.037739 (rank : 29) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.036104 (rank : 30) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
BAALC_MOUSE
|
||||||
| NC score | 0.034091 (rank : 31) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHV1, Q8VBS8, Q9CYS9 | Gene names | Baalc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain and acute leukemia cytoplasmic protein. | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.034009 (rank : 32) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
BLNK_HUMAN
|
||||||
| NC score | 0.033751 (rank : 33) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.033696 (rank : 34) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.032916 (rank : 35) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.032685 (rank : 36) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.032413 (rank : 37) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.032123 (rank : 38) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.032102 (rank : 39) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.032097 (rank : 40) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.031610 (rank : 41) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
CS029_MOUSE
|
||||||
| NC score | 0.031598 (rank : 42) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
INGR1_MOUSE
|
||||||
| NC score | 0.030308 (rank : 43) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15261 | Gene names | Ifngr1, Ifngr | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-gamma receptor alpha chain precursor (IFN-gamma-R1) (CD119 antigen). | |||||
|
MCM3A_MOUSE
|
||||||
| NC score | 0.030035 (rank : 44) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
BNC2_HUMAN
|
||||||
| NC score | 0.029123 (rank : 45) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
BNC2_MOUSE
|
||||||
| NC score | 0.028921 (rank : 46) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.028796 (rank : 47) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CABYR_MOUSE
|
||||||
| NC score | 0.027282 (rank : 48) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.025013 (rank : 49) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.024898 (rank : 50) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
VP13B_HUMAN
|
||||||
| NC score | 0.024552 (rank : 51) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7G8, Q709C6, Q709C7, Q7Z7G4, Q7Z7G5, Q7Z7G6, Q7Z7G7, Q8NB77, Q9NWV1, Q9Y4E7 | Gene names | VPS13B, CHS1, COH1, KIAA0532 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 13B (Cohen syndrome protein 1). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.023801 (rank : 52) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.023801 (rank : 53) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SMAD5_HUMAN
|
||||||
| NC score | 0.023746 (rank : 54) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| NC score | 0.023318 (rank : 55) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.022341 (rank : 56) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.022209 (rank : 57) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.021867 (rank : 58) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.021621 (rank : 59) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.021068 (rank : 60) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.020670 (rank : 61) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
KCT2_MOUSE
|
||||||
| NC score | 0.020187 (rank : 62) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K201, Q5SVC5, Q922L7, Q9CVN1 | Gene names | Kct2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratinocytes-associated transmembrane protein 2 precursor. | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.020047 (rank : 63) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.019812 (rank : 64) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.019718 (rank : 65) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
RIMB2_HUMAN
|
||||||
| NC score | 0.019379 (rank : 66) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
AP180_MOUSE
|
||||||
| NC score | 0.019280 (rank : 67) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.019099 (rank : 68) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
TLE2_MOUSE
|
||||||
| NC score | 0.018508 (rank : 69) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVB2 | Gene names | Tle2, Grg2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.017975 (rank : 70) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
RIMB2_MOUSE
|
||||||
| NC score | 0.017706 (rank : 71) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U40 | Gene names | Rimbp2, Kiaa0318, Rbp2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.017644 (rank : 72) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
BGLR_MOUSE
|
||||||
| NC score | 0.017603 (rank : 73) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12265, Q61601, Q64473, Q64474 | Gene names | Gusb, Gus, Gus-s | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31). | |||||
|
PHF13_MOUSE
|
||||||
| NC score | 0.017572 (rank : 74) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2W6 | Gene names | Phf13 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 13. | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.017511 (rank : 75) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
DBND2_HUMAN
|
||||||
| NC score | 0.017278 (rank : 76) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |

|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.017036 (rank : 77) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.016051 (rank : 78) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.015979 (rank : 79) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.015323 (rank : 80) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
TLE2_HUMAN
|
||||||
| NC score | 0.015145 (rank : 81) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.014857 (rank : 82) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
PJA1_MOUSE
|
||||||
| NC score | 0.014268 (rank : 83) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
CYB5B_MOUSE
|
||||||
| NC score | 0.013756 (rank : 84) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQX2, Q9D1M6, Q9D8R3 | Gene names | Cyb5b, Cyb5m | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytochrome b5 type B precursor (Cytochrome b5 outer mitochondrial membrane isoform). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.012704 (rank : 85) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.012471 (rank : 86) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | 0.012188 (rank : 87) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.012001 (rank : 88) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.011981 (rank : 89) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HIRA_MOUSE
|
||||||
| NC score | 0.011725 (rank : 90) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61666, O08845, Q62365 | Gene names | Hira, Tuple1 | |||
|
Domain Architecture |

|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.011694 (rank : 91) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
GTF2I_MOUSE
|
||||||
| NC score | 0.011277 (rank : 92) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESZ8, O54700, O55030, O55031, Q8VHD1, Q8VHD2, Q8VHD3, Q8VHD4, Q9CSB5, Q9D9K9 | Gene names | Gtf2i, Bap135, Diws1t | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.011225 (rank : 93) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
XYLT1_HUMAN
|
||||||
| NC score | 0.011073 (rank : 94) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y38, Q9H1B6 | Gene names | XYLT1, XT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xylosyltransferase 1 (EC 2.4.2.26) (Xylosyltransferase I) (XylT-I) (XT-I) (Peptide O-xylosyltransferase 1). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.010894 (rank : 95) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.010841 (rank : 96) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.009960 (rank : 97) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
KS6C1_MOUSE
|
||||||
| NC score | 0.009884 (rank : 98) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BLK9, Q3UDY3, Q6PDY4, Q8BMQ5, Q8BX49 | Gene names | Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase). | |||||
|
RPGF2_HUMAN
|
||||||
| NC score | 0.009656 (rank : 99) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.009610 (rank : 100) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
PI3R4_MOUSE
|
||||||
| NC score | 0.008826 (rank : 101) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VD65, Q56A65, Q8C948, Q8C9D7, Q9CVC5 | Gene names | Pik3r4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoinositide 3-kinase regulatory subunit 4 (EC 2.7.11.1) (PI3- kinase regulatory subunit 4). | |||||
|
SUHW4_HUMAN
|
||||||
| NC score | 0.008758 (rank : 102) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
LPIN3_MOUSE
|
||||||
| NC score | 0.008675 (rank : 103) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-3. | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.008666 (rank : 104) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.008247 (rank : 105) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
NFL_HUMAN
|
||||||
| NC score | 0.008113 (rank : 106) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
ZHX3_MOUSE
|
||||||
| NC score | 0.007884 (rank : 107) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0Q2, Q80U14 | Gene names | Zhx3, Kiaa0395, Tix1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and homeodomain protein 3 (Triple homeobox protein 1). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.007805 (rank : 108) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.007628 (rank : 109) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.007501 (rank : 110) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.007321 (rank : 111) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
FGD3_MOUSE
|
||||||
| NC score | 0.006969 (rank : 112) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.005694 (rank : 113) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.005481 (rank : 114) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
ARAF_HUMAN
|
||||||
| NC score | 0.005140 (rank : 115) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10398, P07557, Q5H9B3 | Gene names | ARAF, ARAF1, PKS, PKS2 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (A- raf-1) (Proto-oncogene Pks). | |||||
|
KAISO_HUMAN
|
||||||
| NC score | 0.005051 (rank : 116) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86T24, O00319, Q7Z361, Q8IVP6, Q8N3P0 | Gene names | ZBTB33, KAISO, ZNF348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
SNRK_MOUSE
|
||||||
| NC score | 0.004440 (rank : 117) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 849 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDU5, Q91WX6 | Gene names | Snrk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.003691 (rank : 118) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
SNRK_HUMAN
|
||||||
| NC score | 0.003188 (rank : 119) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRH2, Q14706, Q68D15, Q6IQ46, Q9NXI7 | Gene names | SNRK, KIAA0096, SNFRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
IRAK1_HUMAN
|
||||||
| NC score | 0.002227 (rank : 120) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51617, Q7Z5V4, Q96RL2 | Gene names | IRAK1, IRAK | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1). | |||||
|
ZSCA2_MOUSE
|
||||||
| NC score | -0.001098 (rank : 121) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 121 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||