Please be patient as the page loads
|
SMAD9_MOUSE
|
||||||
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |
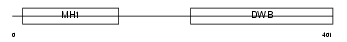
|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMAD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988309 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.988540 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.989779 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.988495 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD9_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.992233 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD9_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD3_HUMAN
|
||||||
| θ value | 3.38586e-148 (rank : 7) | NC score | 0.987819 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |
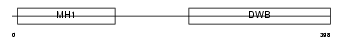
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| θ value | 3.38586e-148 (rank : 8) | NC score | 0.987819 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |
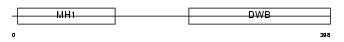
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_HUMAN
|
||||||
| θ value | 7.30591e-143 (rank : 9) | NC score | 0.986648 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD2_MOUSE
|
||||||
| θ value | 1.62758e-142 (rank : 10) | NC score | 0.986646 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 4.38363e-63 (rank : 11) | NC score | 0.914955 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD4_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 12) | NC score | 0.915432 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD6_MOUSE
|
||||||
| θ value | 1.08979e-29 (rank : 13) | NC score | 0.802203 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |
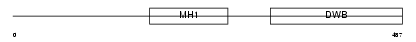
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD6_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 14) | NC score | 0.799259 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD7_HUMAN
|
||||||
| θ value | 9.54697e-26 (rank : 15) | NC score | 0.787064 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
SMAD7_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 16) | NC score | 0.787806 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
BRSK2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.001055 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
ROBO4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.010863 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WZ75, Q8TEG1, Q96JV6, Q9H718, Q9NWJ8 | Gene names | ROBO4 | |||
|
Domain Architecture |
|
|||||
| Description | Roundabout homolog 4 precursor (Magic roundabout). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.021950 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.021844 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.020273 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.011344 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.020048 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.020064 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.010819 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.002157 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.003485 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.017989 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.017891 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.005190 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
B3A3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.006731 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48751 | Gene names | SLC4A3, AE3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3) (Cardiac/brain band 3-like protein) (CAE3/BAE3). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.016859 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GT2D1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.025386 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
PAK7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | -0.000483 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P286, Q5W115, Q9BX09, Q9ULF6 | Gene names | PAK7, KIAA1264, PAK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (PAK-5). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.007560 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.013825 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
KBTB4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.016399 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVX7, Q6IA85, Q9BUC3, Q9NV76 | Gene names | KBTBD4, BKLHD4 | |||
|
Domain Architecture |
|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
RIN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.017527 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
EMID1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.007967 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96A84, Q6ICG1, Q86SS7 | Gene names | EMID1, EMU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
FBX41_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.016292 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
FOXJ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.004052 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92949, O00630 | Gene names | FOXJ1, FKHL13, HFH4 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.008551 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.007094 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.002871 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
FOXP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.004075 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.005280 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MINK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | -0.000958 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.009321 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.011623 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.001056 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.006303 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.008787 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.004779 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
EMID1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.006916 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91VF5 | Gene names | Emid1, Emu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.003468 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
F100A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.008715 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TB05, Q71MF6 | Gene names | FAM100A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100A. | |||||
|
K0310_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.010750 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.006132 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.011024 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.005012 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RFIP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.000758 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.006336 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
TENS4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.015862 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
TNFL6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.007307 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48023, Q9BZP9 | Gene names | FASLG, APT1LG1, FASL, TNFSF6 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor ligand superfamily member 6 (Fas antigen ligand) (Fas ligand) (CD178 antigen) (CD95L protein) (Apoptosis antigen ligand) (APTL) [Contains: Tumor necrosis factor ligand superfamily member 6, membrane form; Tumor necrosis factor ligand superfamily member 6, soluble form]. | |||||
|
CI079_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.009300 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.003878 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
FGFR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | -0.001965 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21802, P18443, Q01742, Q12922, Q14300, Q14301, Q14302, Q14303, Q14304, Q14305, Q14672, Q14718, Q14719, Q86YI4, Q96KL9, Q96KM0, Q96KM1, Q96KM2, Q9NZU2, Q9NZU3, Q9UD01, Q9UD02, Q9UIH3, Q9UIH4, Q9UIH5, Q9UIH6, Q9UIH7, Q9UIH8, Q9UM87, Q9UMC6, Q9UNS7, Q9UQH7, Q9UQH8, Q9UQH9, Q9UQI0 | Gene names | FGFR2, BEK, KSAM | |||
|
Domain Architecture |

|
|||||
| Description | Fibroblast growth factor receptor 2 precursor (EC 2.7.10.1) (FGFR-2) (Keratinocyte growth factor receptor 2) (CD332 antigen). | |||||
|
FYB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.012920 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.001597 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
LEG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.004151 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16110 | Gene names | Lgals3 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-34 galactoside-binding lectin). | |||||
|
NAT6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.008974 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R123, Q8QZY5, Q8R014, Q9ERM0 | Gene names | Nat6, Fus2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
NDST2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.002525 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52849, Q2TB32, Q59H89 | Gene names | NDST2, HSST2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.003734 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
WNT16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.005281 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYS1 | Gene names | Wnt16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-16 precursor. | |||||
|
ZN645_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.019808 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.074517 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
SMAD9_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD9_HUMAN
|
||||||
| NC score | 0.992233 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD5_HUMAN
|
||||||
| NC score | 0.989779 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD1_MOUSE
|
||||||
| NC score | 0.988540 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD5_MOUSE
|
||||||
| NC score | 0.988495 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD1_HUMAN
|
||||||
| NC score | 0.988309 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD3_HUMAN
|
||||||
| NC score | 0.987819 (rank : 7) | θ value | 3.38586e-148 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| NC score | 0.987819 (rank : 8) | θ value | 3.38586e-148 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_HUMAN
|
||||||
| NC score | 0.986648 (rank : 9) | θ value | 7.30591e-143 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD2_MOUSE
|
||||||
| NC score | 0.986646 (rank : 10) | θ value | 1.62758e-142 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD4_HUMAN
|
||||||
| NC score | 0.915432 (rank : 11) | θ value | 4.88048e-38 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.914955 (rank : 12) | θ value | 4.38363e-63 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD6_MOUSE
|
||||||
| NC score | 0.802203 (rank : 13) | θ value | 1.08979e-29 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD6_HUMAN
|
||||||
| NC score | 0.799259 (rank : 14) | θ value | 2.05525e-28 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD7_MOUSE
|
||||||
| NC score | 0.787806 (rank : 15) | θ value | 1.24688e-25 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
SMAD7_HUMAN
|
||||||
| NC score | 0.787064 (rank : 16) | θ value | 9.54697e-26 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.074517 (rank : 17) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
GT2D1_MOUSE
|
||||||
| NC score | 0.025386 (rank : 18) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.021950 (rank : 19) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.021844 (rank : 20) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.020273 (rank : 21) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.020064 (rank : 22) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.020048 (rank : 23) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
ZN645_HUMAN
|
||||||
| NC score | 0.019808 (rank : 24) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.017989 (rank : 25) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.017891 (rank : 26) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RIN3_HUMAN
|
||||||
| NC score | 0.017527 (rank : 27) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.016859 (rank : 28) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
KBTB4_HUMAN
|
||||||
| NC score | 0.016399 (rank : 29) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVX7, Q6IA85, Q9BUC3, Q9NV76 | Gene names | KBTBD4, BKLHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
FBX41_MOUSE
|
||||||
| NC score | 0.016292 (rank : 30) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.015862 (rank : 31) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.013825 (rank : 32) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.012920 (rank : 33) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.011623 (rank : 34) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.011344 (rank : 35) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.011024 (rank : 36) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
ROBO4_HUMAN
|
||||||
| NC score | 0.010863 (rank : 37) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WZ75, Q8TEG1, Q96JV6, Q9H718, Q9NWJ8 | Gene names | ROBO4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor (Magic roundabout). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.010819 (rank : 38) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.010750 (rank : 39) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.009321 (rank : 40) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.009300 (rank : 41) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
NAT6_MOUSE
|
||||||
| NC score | 0.008974 (rank : 42) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R123, Q8QZY5, Q8R014, Q9ERM0 | Gene names | Nat6, Fus2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.008787 (rank : 43) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
F100A_HUMAN
|
||||||
| NC score | 0.008715 (rank : 44) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TB05, Q71MF6 | Gene names | FAM100A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100A. | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.008551 (rank : 45) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
EMID1_HUMAN
|
||||||
| NC score | 0.007967 (rank : 46) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96A84, Q6ICG1, Q86SS7 | Gene names | EMID1, EMU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.007560 (rank : 47) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
TNFL6_HUMAN
|
||||||
| NC score | 0.007307 (rank : 48) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48023, Q9BZP9 | Gene names | FASLG, APT1LG1, FASL, TNFSF6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor ligand superfamily member 6 (Fas antigen ligand) (Fas ligand) (CD178 antigen) (CD95L protein) (Apoptosis antigen ligand) (APTL) [Contains: Tumor necrosis factor ligand superfamily member 6, membrane form; Tumor necrosis factor ligand superfamily member 6, soluble form]. | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.007094 (rank : 49) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
EMID1_MOUSE
|
||||||
| NC score | 0.006916 (rank : 50) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91VF5 | Gene names | Emid1, Emu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
B3A3_HUMAN
|
||||||
| NC score | 0.006731 (rank : 51) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48751 | Gene names | SLC4A3, AE3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3) (Cardiac/brain band 3-like protein) (CAE3/BAE3). | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.006336 (rank : 52) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.006303 (rank : 53) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.006132 (rank : 54) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
WNT16_MOUSE
|
||||||
| NC score | 0.005281 (rank : 55) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYS1 | Gene names | Wnt16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-16 precursor. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.005280 (rank : 56) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.005190 (rank : 57) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.005012 (rank : 58) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.004779 (rank : 59) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
LEG3_MOUSE
|
||||||
| NC score | 0.004151 (rank : 60) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16110 | Gene names | Lgals3 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-34 galactoside-binding lectin). | |||||
|
FOXP4_MOUSE
|
||||||
| NC score | 0.004075 (rank : 61) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
FOXJ1_HUMAN
|
||||||
| NC score | 0.004052 (rank : 62) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92949, O00630 | Gene names | FOXJ1, FKHL13, HFH4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.003878 (rank : 63) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.003734 (rank : 64) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.003485 (rank : 65) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.003468 (rank : 66) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.002871 (rank : 67) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
NDST2_HUMAN
|
||||||
| NC score | 0.002525 (rank : 68) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52849, Q2TB32, Q59H89 | Gene names | NDST2, HSST2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 2 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 2) (NDST- 2) (N-heparan sulfate sulfotransferase 2) (N-HSST 2) [Includes: Heparan sulfate N-deacetylase 2 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 2 (EC 2.8.2.-)]. | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.002157 (rank : 69) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.001597 (rank : 70) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
PCD15_HUMAN
|
||||||
| NC score | 0.001056 (rank : 71) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
BRSK2_HUMAN
|
||||||
| NC score | 0.001055 (rank : 72) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
RFIP4_HUMAN
|
||||||
| NC score | 0.000758 (rank : 73) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
PAK7_HUMAN
|
||||||
| NC score | -0.000483 (rank : 74) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P286, Q5W115, Q9BX09, Q9ULF6 | Gene names | PAK7, KIAA1264, PAK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 7 (EC 2.7.11.1) (p21-activated kinase 7) (PAK-7) (PAK-5). | |||||
|
MINK1_HUMAN
|
||||||
| NC score | -0.000958 (rank : 75) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
FGFR2_HUMAN
|
||||||
| NC score | -0.001965 (rank : 76) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21802, P18443, Q01742, Q12922, Q14300, Q14301, Q14302, Q14303, Q14304, Q14305, Q14672, Q14718, Q14719, Q86YI4, Q96KL9, Q96KM0, Q96KM1, Q96KM2, Q9NZU2, Q9NZU3, Q9UD01, Q9UD02, Q9UIH3, Q9UIH4, Q9UIH5, Q9UIH6, Q9UIH7, Q9UIH8, Q9UM87, Q9UMC6, Q9UNS7, Q9UQH7, Q9UQH8, Q9UQH9, Q9UQI0 | Gene names | FGFR2, BEK, KSAM | |||
|
Domain Architecture |

|
|||||
| Description | Fibroblast growth factor receptor 2 precursor (EC 2.7.10.1) (FGFR-2) (Keratinocyte growth factor receptor 2) (CD332 antigen). | |||||