Please be patient as the page loads
|
SMAD9_HUMAN
|
||||||
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMAD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992661 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992606 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.994002 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.992519 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD9_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD9_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.992233 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |
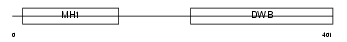
|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD3_HUMAN
|
||||||
| θ value | 2.2711e-144 (rank : 7) | NC score | 0.987564 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |
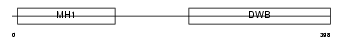
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| θ value | 2.2711e-144 (rank : 8) | NC score | 0.987564 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |
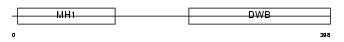
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_HUMAN
|
||||||
| θ value | 6.40027e-139 (rank : 9) | NC score | 0.984538 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD2_MOUSE
|
||||||
| θ value | 1.42583e-138 (rank : 10) | NC score | 0.984535 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 1.00825e-67 (rank : 11) | NC score | 0.925348 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD4_HUMAN
|
||||||
| θ value | 2.4834e-66 (rank : 12) | NC score | 0.926339 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD6_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 13) | NC score | 0.775129 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |
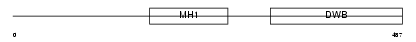
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD6_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 14) | NC score | 0.772192 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |
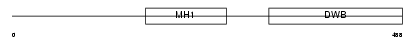
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD7_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 15) | NC score | 0.759643 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |
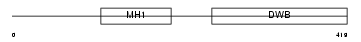
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
SMAD7_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 16) | NC score | 0.760462 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |
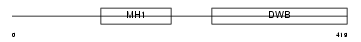
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.031489 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
HCLS1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.015990 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.020292 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.034302 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.019978 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
CC024_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.028646 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PS1 | Gene names | C3orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf24. | |||||
|
MLTK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | -0.000863 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.033487 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.043445 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
MYO1E_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.004263 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12965, Q14778 | Gene names | MYO1E, MYO1C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ie (Myosin Ic). | |||||
|
US6NL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.010962 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
FOXJ1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.016414 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |
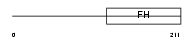
|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.039390 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.016720 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
GT2D1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.025599 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
KI3L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.009427 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43629, O43473, Q16541 | Gene names | KIR3DL1, NKAT3, NKB1 | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 3) (NKAT-3) (p70 natural killer cell receptor clones CL-2/CL-11) (HLA-BW4-specific inhibitory NK cell receptor). | |||||
|
KI3L3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.009568 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N743, O95056, Q7Z7N4, Q8NHI2, Q8NHI3, Q9UEI4 | Gene names | KIR3DL3, CD158Z, KIR3DL7, KIRC1 | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL3 precursor (Killer cell inhibitory receptor 1) (CD158z antigen). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.005256 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.005944 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
ZBT38_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | -0.002180 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.011803 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
KI3S1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.008586 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14943 | Gene names | KIR3DS1, NKAT10 | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DS1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 10) (NKAT-10). | |||||
|
ARHG7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.003708 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
CC024_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.020752 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D4K4, Q80XT0, Q80Y40 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf24 homolog. | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.006265 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.006172 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | -0.001886 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SIGL5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.003396 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920G3 | Gene names | Siglec5, Siglecf | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 5 precursor (Siglec-5) (Sialic acid-binding Ig-like lectin-F) (mSiglec-F). | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.002727 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | -0.000276 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SIG10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.002372 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
WNT16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.005388 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYS1 | Gene names | Wnt16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-16 precursor. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.071991 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
SMAD9_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD5_HUMAN
|
||||||
| NC score | 0.994002 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD1_HUMAN
|
||||||
| NC score | 0.992661 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD1_MOUSE
|
||||||
| NC score | 0.992606 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD5_MOUSE
|
||||||
| NC score | 0.992519 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD9_MOUSE
|
||||||
| NC score | 0.992233 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD3_HUMAN
|
||||||
| NC score | 0.987564 (rank : 7) | θ value | 2.2711e-144 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| NC score | 0.987564 (rank : 8) | θ value | 2.2711e-144 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_HUMAN
|
||||||
| NC score | 0.984538 (rank : 9) | θ value | 6.40027e-139 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD2_MOUSE
|
||||||
| NC score | 0.984535 (rank : 10) | θ value | 1.42583e-138 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD4_HUMAN
|
||||||
| NC score | 0.926339 (rank : 11) | θ value | 2.4834e-66 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.925348 (rank : 12) | θ value | 1.00825e-67 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD6_MOUSE
|
||||||
| NC score | 0.775129 (rank : 13) | θ value | 4.58923e-20 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD6_HUMAN
|
||||||
| NC score | 0.772192 (rank : 14) | θ value | 2.97466e-19 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD7_MOUSE
|
||||||
| NC score | 0.760462 (rank : 15) | θ value | 2.5182e-18 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
SMAD7_HUMAN
|
||||||
| NC score | 0.759643 (rank : 16) | θ value | 2.5182e-18 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.071991 (rank : 17) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.043445 (rank : 18) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.039390 (rank : 19) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.034302 (rank : 20) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.033487 (rank : 21) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.031489 (rank : 22) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
CC024_HUMAN
|
||||||
| NC score | 0.028646 (rank : 23) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PS1 | Gene names | C3orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf24. | |||||
|
GT2D1_MOUSE
|
||||||
| NC score | 0.025599 (rank : 24) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
CC024_MOUSE
|
||||||
| NC score | 0.020752 (rank : 25) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D4K4, Q80XT0, Q80Y40 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf24 homolog. | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.020292 (rank : 26) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.019978 (rank : 27) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.016720 (rank : 28) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
FOXJ1_MOUSE
|
||||||
| NC score | 0.016414 (rank : 29) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
HCLS1_HUMAN
|
||||||
| NC score | 0.015990 (rank : 30) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.011803 (rank : 31) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
US6NL_MOUSE
|
||||||
| NC score | 0.010962 (rank : 32) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
KI3L3_HUMAN
|
||||||
| NC score | 0.009568 (rank : 33) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N743, O95056, Q7Z7N4, Q8NHI2, Q8NHI3, Q9UEI4 | Gene names | KIR3DL3, CD158Z, KIR3DL7, KIRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL3 precursor (Killer cell inhibitory receptor 1) (CD158z antigen). | |||||
|
KI3L1_HUMAN
|
||||||
| NC score | 0.009427 (rank : 34) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43629, O43473, Q16541 | Gene names | KIR3DL1, NKAT3, NKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 3) (NKAT-3) (p70 natural killer cell receptor clones CL-2/CL-11) (HLA-BW4-specific inhibitory NK cell receptor). | |||||
|
KI3S1_HUMAN
|
||||||
| NC score | 0.008586 (rank : 35) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14943 | Gene names | KIR3DS1, NKAT10 | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 3DS1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 10) (NKAT-10). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.006265 (rank : 36) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.006172 (rank : 37) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.005944 (rank : 38) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
WNT16_MOUSE
|
||||||
| NC score | 0.005388 (rank : 39) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYS1 | Gene names | Wnt16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-16 precursor. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.005256 (rank : 40) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYO1E_HUMAN
|
||||||
| NC score | 0.004263 (rank : 41) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12965, Q14778 | Gene names | MYO1E, MYO1C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ie (Myosin Ic). | |||||
|
ARHG7_HUMAN
|
||||||
| NC score | 0.003708 (rank : 42) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
SIGL5_MOUSE
|
||||||
| NC score | 0.003396 (rank : 43) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920G3 | Gene names | Siglec5, Siglecf | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 5 precursor (Siglec-5) (Sialic acid-binding Ig-like lectin-F) (mSiglec-F). | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.002727 (rank : 44) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
SIG10_HUMAN
|
||||||
| NC score | 0.002372 (rank : 45) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | -0.000276 (rank : 46) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
MLTK_HUMAN
|
||||||
| NC score | -0.000863 (rank : 47) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYL2, Q53SX1, Q580W8, Q59GY5, Q86YW8, Q9HCC4, Q9HCC5, Q9HDD2, Q9NYE9 | Gene names | MLTK, ZAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase MLT (EC 2.7.11.25) (MLK-like mitogen-activated protein triple kinase) (Leucine zipper- and sterile alpha motif-containing kinase) (Sterile alpha motif- and leucine zipper-containing kinase AZK) (Mixed lineage kinase-related kinase) (MLK-related kinase) (MRK) (Cervical cancer suppressor gene 4 protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | -0.001886 (rank : 48) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZBT38_HUMAN
|
||||||
| NC score | -0.002180 (rank : 49) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||