Please be patient as the page loads
|
NFAC3_MOUSE
|
||||||
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NFAC3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989556 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC1_MOUSE
|
||||||
| θ value | 9.54178e-143 (rank : 3) | NC score | 0.949773 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 1.37783e-141 (rank : 4) | NC score | 0.938746 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 5.23575e-141 (rank : 5) | NC score | 0.917271 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC2_MOUSE
|
||||||
| θ value | 4.4633e-116 (rank : 6) | NC score | 0.930662 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | 2.44908e-114 (rank : 7) | NC score | 0.926087 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 1.61343e-57 (rank : 8) | NC score | 0.809778 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | 1.61343e-57 (rank : 9) | NC score | 0.829864 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
TF65_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 10) | NC score | 0.265367 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TF65_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 11) | NC score | 0.253484 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
NFKB2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 12) | NC score | 0.112063 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 13) | NC score | 0.105766 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.057696 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.036084 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.031882 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.071972 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.049096 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.027391 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.058792 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
K1683_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.071929 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
SMAD9_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.020292 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.034372 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.030194 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
MUC15_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.039143 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N387, Q6UWS3, Q8IXI8, Q8WW41 | Gene names | MUC15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.043478 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
DCX_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.036024 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.046433 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.012699 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.022355 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.020927 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RBMS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.016526 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
RIPK3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.007105 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |
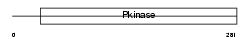
|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.004704 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
ULK2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.004726 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.041909 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.030569 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
GA2L3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.021895 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.017515 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.006381 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.011547 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
BCL7B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.027019 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQE9, O43769, Q13845, Q6ZW75 | Gene names | BCL7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
BCL7B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.027027 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921K9, O89022, Q3TV31, Q3U2W0 | Gene names | Bcl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.027153 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.027770 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
DCX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.028808 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |
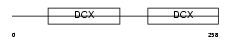
|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.030739 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
SAM10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.056192 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYL1 | Gene names | SAMD10, C20orf136 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
SAM10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.055128 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TST3 | Gene names | Samd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.017756 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
YETS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.022909 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.002807 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.013886 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
DLX5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.006261 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
NU214_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.031012 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.023229 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.004102 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BRAC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.007693 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20293 | Gene names | T | |||
|
Domain Architecture |

|
|||||
| Description | Brachyury protein (T protein). | |||||
|
DCR1C_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.015343 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4J0, Q8BG72, Q8BTT1 | Gene names | Dclre1c, Art, Snm1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemis protein (EC 3.1.-.-) (DNA cross-link repair 1C protein) (mArt protein) (SNM1-like protein). | |||||
|
DLX5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.006124 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56178, Q9UPL1 | Gene names | DLX5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.004566 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.012137 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
POF1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.013671 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.001385 (rank : 110) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.028421 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.047783 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
I18RA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.012687 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2B1 | Gene names | Il18rap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-18 receptor accessory protein precursor (IL-18 receptor accessory protein) (IL-18RAcP) (Interleukin-18 receptor accessory protein-like) (IL-18Rbeta) (IL-1R accessory protein-like) (IL-1RAcPL) (Accessory protein-like) (AcPL) (IL-1R7). | |||||
|
MDFI_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.023376 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70331, P70330, P70332, Q99JM9 | Gene names | Mdfi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MyoD family inhibitor (Myogenic repressor I-mf). | |||||
|
NEK4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.001579 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1J2, O35673, Q9R1J1 | Gene names | Nek4, Stk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2). | |||||
|
PTC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.014319 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
RBMS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.013348 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |
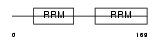
|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.014022 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
IC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.004260 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
PPR3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.012483 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.017431 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.007570 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.022583 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TENS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.017898 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
THB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.002647 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37244 | Gene names | Thrb, C-erba-beta, Nr1a2 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-2. | |||||
|
TTLL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.019266 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.004898 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.019443 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.050770 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CK024_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.038263 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CN037_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.013550 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.022037 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.022429 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.007688 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
KLK6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.000396 (rank : 111) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92876 | Gene names | KLK6, PRSS18, PRSS9 | |||
|
Domain Architecture |
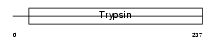
|
|||||
| Description | Kallikrein-6 precursor (EC 3.4.21.-) (Protease M) (Neurosin) (Zyme) (SP59). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.002073 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.014213 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.002709 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ATS19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.001432 (rank : 109) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59509 | Gene names | Adamts19 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 19) (ADAM-TS 19) (ADAM-TS19). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.001969 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.004959 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CT151_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.014158 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |
|
|||||
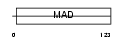
| Description | Uncharacterized protein C20orf151. | |||||
|
EGLN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.028424 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63961, Q61520 | Gene names | Eng, Edg | |||
|
Domain Architecture |

|
|||||
| Description | Endoglin precursor (Cell surface MJ7/18 antigen). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.007182 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
FOLH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.004704 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35409, Q9DCC2 | Gene names | Folh1, Mopsm, Naalad1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate carboxypeptidase 2 (EC 3.4.17.21) (Glutamate carboxypeptidase II) (Membrane glutamate carboxypeptidase) (mGCP) (N- acetylated-alpha-linked acidic dipeptidase I) (NAALADase I) (Pteroylpoly-gamma-glutamate carboxypeptidase) (Folylpoly-gamma- glutamate carboxypeptidase) (FGCP) (Folate hydrolase 1) (Prostate- specific membrane antigen homolog). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.010774 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.015712 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.044981 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.012304 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.022541 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TBX6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.005289 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95947, Q8TAS4 | Gene names | TBX6 | |||
|
Domain Architecture |
|
|||||
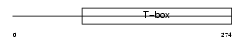
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
TRIP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.015380 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
NBR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.051043 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15453 | Gene names | NBR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NBR2 (Next to BRCA1 gene 2 protein). | |||||
|
RELB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.100587 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |
|
|||||
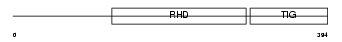
| Description | Transcription factor RelB (I-Rel). | |||||
|
RELB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.097202 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |
|
|||||
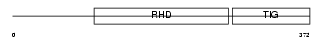
| Description | Transcription factor RelB. | |||||
|
REL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.084363 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
REL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.072462 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.989556 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC1_MOUSE
|
||||||
| NC score | 0.949773 (rank : 3) | θ value | 9.54178e-143 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.938746 (rank : 4) | θ value | 1.37783e-141 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NFAC2_MOUSE
|
||||||
| NC score | 0.930662 (rank : 5) | θ value | 4.4633e-116 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.926087 (rank : 6) | θ value | 2.44908e-114 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.917271 (rank : 7) | θ value | 5.23575e-141 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.829864 (rank : 8) | θ value | 1.61343e-57 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.809778 (rank : 9) | θ value | 1.61343e-57 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
TF65_HUMAN
|
||||||
| NC score | 0.265367 (rank : 10) | θ value | 3.64472e-09 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TF65_MOUSE
|
||||||
| NC score | 0.253484 (rank : 11) | θ value | 1.17247e-07 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
NFKB2_MOUSE
|
||||||
| NC score | 0.112063 (rank : 12) | θ value | 0.000461057 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB2_HUMAN
|
||||||
| NC score | 0.105766 (rank : 13) | θ value | 0.00228821 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
RELB_HUMAN
|
||||||
| NC score | 0.100587 (rank : 14) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB (I-Rel). | |||||
|
RELB_MOUSE
|
||||||
| NC score | 0.097202 (rank : 15) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB. | |||||
|
REL_HUMAN
|
||||||
| NC score | 0.084363 (rank : 16) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.072462 (rank : 17) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.071972 (rank : 18) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.071929 (rank : 19) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.058792 (rank : 20) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.057696 (rank : 21) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SAM10_HUMAN
|
||||||
| NC score | 0.056192 (rank : 22) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYL1 | Gene names | SAMD10, C20orf136 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
SAM10_MOUSE
|
||||||
| NC score | 0.055128 (rank : 23) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TST3 | Gene names | Samd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
NBR2_HUMAN
|
||||||
| NC score | 0.051043 (rank : 24) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15453 | Gene names | NBR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NBR2 (Next to BRCA1 gene 2 protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.050770 (rank : 25) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.049096 (rank : 26) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.047783 (rank : 27) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.046433 (rank : 28) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.044981 (rank : 29) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.043478 (rank : 30) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.041909 (rank : 31) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MUC15_HUMAN
|
||||||
| NC score | 0.039143 (rank : 32) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N387, Q6UWS3, Q8IXI8, Q8WW41 | Gene names | MUC15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.038263 (rank : 33) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.036084 (rank : 34) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.036024 (rank : 35) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.034372 (rank : 36) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.031882 (rank : 37) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.031012 (rank : 38) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.030739 (rank : 39) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.030569 (rank : 40) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.030194 (rank : 41) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
DCX_HUMAN
|
||||||
| NC score | 0.028808 (rank : 42) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
EGLN_MOUSE
|
||||||
| NC score | 0.028424 (rank : 43) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63961, Q61520 | Gene names | Eng, Edg | |||
|
Domain Architecture |

|
|||||
| Description | Endoglin precursor (Cell surface MJ7/18 antigen). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.028421 (rank : 44) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.027770 (rank : 45) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.027391 (rank : 46) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.027153 (rank : 47) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
BCL7B_MOUSE
|
||||||
| NC score | 0.027027 (rank : 48) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921K9, O89022, Q3TV31, Q3U2W0 | Gene names | Bcl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
BCL7B_HUMAN
|
||||||
| NC score | 0.027019 (rank : 49) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQE9, O43769, Q13845, Q6ZW75 | Gene names | BCL7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member B. | |||||
|
MDFI_MOUSE
|
||||||
| NC score | 0.023376 (rank : 50) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70331, P70330, P70332, Q99JM9 | Gene names | Mdfi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MyoD family inhibitor (Myogenic repressor I-mf). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.023229 (rank : 51) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
YETS2_MOUSE
|
||||||
| NC score | 0.022909 (rank : 52) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.022583 (rank : 53) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.022541 (rank : 54) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
EYA4_MOUSE
|
||||||
| NC score | 0.022429 (rank : 55) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.022355 (rank : 56) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.022037 (rank : 57) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
GA2L3_HUMAN
|
||||||
| NC score | 0.021895 (rank : 58) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.020927 (rank : 59) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SMAD9_HUMAN
|
||||||
| NC score | 0.020292 (rank : 60) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.019443 (rank : 61) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.019266 (rank : 62) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
TENS4_MOUSE
|
||||||
| NC score | 0.017898 (rank : 63) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.017756 (rank : 64) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.017515 (rank : 65) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.017431 (rank : 66) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
RBMS1_MOUSE
|
||||||
| NC score | 0.016526 (rank : 67) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
LPP_MOUSE
|
||||||
| NC score | 0.015712 (rank : 68) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
TRIP6_HUMAN
|
||||||
| NC score | 0.015380 (rank : 69) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
DCR1C_MOUSE
|
||||||
| NC score | 0.015343 (rank : 70) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4J0, Q8BG72, Q8BTT1 | Gene names | Dclre1c, Art, Snm1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemis protein (EC 3.1.-.-) (DNA cross-link repair 1C protein) (mArt protein) (SNM1-like protein). | |||||
|
PTC1_MOUSE
|
||||||
| NC score | 0.014319 (rank : 71) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.014213 (rank : 72) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
CT151_HUMAN
|
||||||
| NC score | 0.014158 (rank : 73) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.014022 (rank : 74) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.013886 (rank : 75) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
POF1B_HUMAN
|
||||||
| NC score | 0.013671 (rank : 76) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.013550 (rank : 77) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
RBMS1_HUMAN
|
||||||
| NC score | 0.013348 (rank : 78) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.012699 (rank : 79) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
I18RA_MOUSE
|
||||||
| NC score | 0.012687 (rank : 80) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2B1 | Gene names | Il18rap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-18 receptor accessory protein precursor (IL-18 receptor accessory protein) (IL-18RAcP) (Interleukin-18 receptor accessory protein-like) (IL-18Rbeta) (IL-1R accessory protein-like) (IL-1RAcPL) (Accessory protein-like) (AcPL) (IL-1R7). | |||||
|
PPR3A_MOUSE
|
||||||
| NC score | 0.012483 (rank : 81) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.012304 (rank : 82) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.012137 (rank : 83) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.011547 (rank : 84) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.010774 (rank : 85) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
BRAC_MOUSE
|
||||||
| NC score | 0.007693 (rank : 86) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20293 | Gene names | T | |||
|
Domain Architecture |

|
|||||
| Description | Brachyury protein (T protein). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.007688 (rank : 87) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.007570 (rank : 88) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.007182 (rank : 89) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
RIPK3_MOUSE
|
||||||
| NC score | 0.007105 (rank : 90) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.006381 (rank : 91) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
DLX5_MOUSE
|
||||||
| NC score | 0.006261 (rank : 92) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
DLX5_HUMAN
|
||||||
| NC score | 0.006124 (rank : 93) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56178, Q9UPL1 | Gene names | DLX5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
TBX6_HUMAN
|
||||||
| NC score | 0.005289 (rank : 94) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95947, Q8TAS4 | Gene names | TBX6 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX6 (T-box protein 6). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.004959 (rank : 95) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | 0.004898 (rank : 96) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
ULK2_MOUSE
|
||||||
| NC score | 0.004726 (rank : 97) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
ULK2_HUMAN
|
||||||
| NC score | 0.004704 (rank : 98) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
FOLH1_MOUSE
|
||||||
| NC score | 0.004704 (rank : 99) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35409, Q9DCC2 | Gene names | Folh1, Mopsm, Naalad1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate carboxypeptidase 2 (EC 3.4.17.21) (Glutamate carboxypeptidase II) (Membrane glutamate carboxypeptidase) (mGCP) (N- acetylated-alpha-linked acidic dipeptidase I) (NAALADase I) (Pteroylpoly-gamma-glutamate carboxypeptidase) (Folylpoly-gamma- glutamate carboxypeptidase) (FGCP) (Folate hydrolase 1) (Prostate- specific membrane antigen homolog). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.004566 (rank : 100) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
IC1_MOUSE
|
||||||
| NC score | 0.004260 (rank : 101) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.004102 (rank : 102) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.002807 (rank : 103) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.002709 (rank : 104) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
THB2_MOUSE
|
||||||
| NC score | 0.002647 (rank : 105) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37244 | Gene names | Thrb, C-erba-beta, Nr1a2 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-2. | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.002073 (rank : 106) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.001969 (rank : 107) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
NEK4_MOUSE
|
||||||
| NC score | 0.001579 (rank : 108) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1J2, O35673, Q9R1J1 | Gene names | Nek4, Stk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2). | |||||
|
ATS19_MOUSE
|
||||||
| NC score | 0.001432 (rank : 109) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59509 | Gene names | Adamts19 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 19) (ADAM-TS 19) (ADAM-TS19). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.001385 (rank : 110) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
KLK6_HUMAN
|
||||||
| NC score | 0.000396 (rank : 111) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92876 | Gene names | KLK6, PRSS18, PRSS9 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-6 precursor (EC 3.4.21.-) (Protease M) (Neurosin) (Zyme) (SP59). | |||||