Please be patient as the page loads
|
EYA4_MOUSE
|
||||||
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EYA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.975708 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.974209 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.991348 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA4_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA2_MOUSE
|
||||||
| θ value | 2.76339e-158 (rank : 5) | NC score | 0.980370 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_HUMAN
|
||||||
| θ value | 2.33935e-157 (rank : 6) | NC score | 0.979375 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |
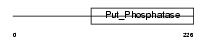
|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA3_HUMAN
|
||||||
| θ value | 2.42085e-154 (rank : 7) | NC score | 0.977207 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA3_MOUSE
|
||||||
| θ value | 4.13891e-146 (rank : 8) | NC score | 0.977410 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 9) | NC score | 0.090358 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 10) | NC score | 0.090345 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.041184 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.059023 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
NFIC_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.035614 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08651, P08652, Q14932, Q9UPJ3, Q9UPJ9, Q9UPK0, Q9UPK1 | Gene names | NFIC, NFI | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.043167 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.057546 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
TFG_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.084474 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.069923 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
EWS_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.117016 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
HNRL1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.068453 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
PEPC_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.029996 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20142, Q5T3D7, Q5T3D8 | Gene names | PGC | |||
|
Domain Architecture |
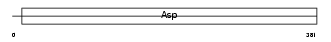
|
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.054613 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.056593 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.030145 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.025801 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
FUS_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.122563 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.017237 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.038204 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MCM3A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.058931 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
PHC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.029442 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
EWS_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.116616 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.006072 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
LEG3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.028826 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |
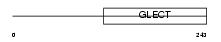
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
MCM3A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.068516 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
NUP53_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.036476 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.024060 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.010331 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.007601 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
FUS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.111984 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.020083 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
NDF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.017791 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.019557 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.019482 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
ERRFI_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.022467 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
HXB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.009095 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P14653 | Gene names | HOXB1, HOX2I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B1 (Hox-2I). | |||||
|
K1718_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.016418 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.018226 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.011559 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
WBP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.026007 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969T9, O95638 | Gene names | WBP2 | |||
|
Domain Architecture |
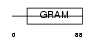
|
|||||
| Description | WW domain-binding protein 2 (WBP-2). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.031992 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.017062 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
K1718_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.014504 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.028371 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.028876 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.001038 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.011015 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.028399 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
DLX4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.004027 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92988, O60480, Q13265, Q6PJK0, Q9HBE0 | Gene names | DLX4, BP1, DLX7, DLX8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein DLX-4 (DLX-7) (DLX-8) (Beta protein 1). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.003141 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.010403 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.022429 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
PMVK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.017111 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1G2, Q8R021, Q9D6K3 | Gene names | Pmvk | |||
|
Domain Architecture |

|
|||||
| Description | Phosphomevalonate kinase (EC 2.7.4.2) (PMKase). | |||||
|
RAB6C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.002161 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0N0, Q9P128 | Gene names | RAB6C, WTH3 | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-6C (Rab6-like protein WTH3). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.012426 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | -0.001916 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.000981 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.034307 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.003050 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DIP2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.012601 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
DIP2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.012679 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
HGFA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.000276 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
ILF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.027552 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
NDF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.011010 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
PMVK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.024602 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15126 | Gene names | PMVK, PMKI | |||
|
Domain Architecture |

|
|||||
| Description | Phosphomevalonate kinase (EC 2.7.4.2) (PMKase). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.017024 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
EYA4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.991348 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA2_MOUSE
|
||||||
| NC score | 0.980370 (rank : 3) | θ value | 2.76339e-158 (rank : 5) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_HUMAN
|
||||||
| NC score | 0.979375 (rank : 4) | θ value | 2.33935e-157 (rank : 6) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA3_MOUSE
|
||||||
| NC score | 0.977410 (rank : 5) | θ value | 4.13891e-146 (rank : 8) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA3_HUMAN
|
||||||
| NC score | 0.977207 (rank : 6) | θ value | 2.42085e-154 (rank : 7) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA1_HUMAN
|
||||||
| NC score | 0.975708 (rank : 7) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA1_MOUSE
|
||||||
| NC score | 0.974209 (rank : 8) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.122563 (rank : 9) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.117016 (rank : 10) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.116616 (rank : 11) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.111984 (rank : 12) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.090358 (rank : 13) | θ value | 0.00390308 (rank : 9) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.090345 (rank : 14) | θ value | 0.00390308 (rank : 10) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
TFG_HUMAN
|
||||||
| NC score | 0.084474 (rank : 15) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.069923 (rank : 16) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
MCM3A_MOUSE
|
||||||
| NC score | 0.068516 (rank : 17) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
HNRL1_HUMAN
|
||||||
| NC score | 0.068453 (rank : 18) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.059023 (rank : 19) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MCM3A_HUMAN
|
||||||
| NC score | 0.058931 (rank : 20) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.057546 (rank : 21) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.056593 (rank : 22) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.054613 (rank : 23) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.043167 (rank : 24) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.041184 (rank : 25) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.038204 (rank : 26) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
NUP53_MOUSE
|
||||||
| NC score | 0.036476 (rank : 27) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
NFIC_HUMAN
|
||||||
| NC score | 0.035614 (rank : 28) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08651, P08652, Q14932, Q9UPJ3, Q9UPJ9, Q9UPK0, Q9UPK1 | Gene names | NFIC, NFI | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.034307 (rank : 29) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.031992 (rank : 30) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.030145 (rank : 31) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
PEPC_HUMAN
|
||||||
| NC score | 0.029996 (rank : 32) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20142, Q5T3D7, Q5T3D8 | Gene names | PGC | |||
|
Domain Architecture |

|
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
PHC1_HUMAN
|
||||||
| NC score | 0.029442 (rank : 33) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.028876 (rank : 34) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
LEG3_HUMAN
|
||||||
| NC score | 0.028826 (rank : 35) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.028399 (rank : 36) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.028371 (rank : 37) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
ILF3_MOUSE
|
||||||
| NC score | 0.027552 (rank : 38) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1X4, Q80VD5, Q812A1, Q8BP80, Q8C2H8, Q8K588 | Gene names | Ilf3 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3. | |||||
|
WBP2_HUMAN
|
||||||
| NC score | 0.026007 (rank : 39) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969T9, O95638 | Gene names | WBP2 | |||
|
Domain Architecture |

|
|||||
| Description | WW domain-binding protein 2 (WBP-2). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.025801 (rank : 40) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PMVK_HUMAN
|
||||||
| NC score | 0.024602 (rank : 41) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15126 | Gene names | PMVK, PMKI | |||
|
Domain Architecture |

|
|||||
| Description | Phosphomevalonate kinase (EC 2.7.4.2) (PMKase). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.024060 (rank : 42) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ERRFI_MOUSE
|
||||||
| NC score | 0.022467 (rank : 43) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.022429 (rank : 44) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.020083 (rank : 45) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.019557 (rank : 46) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.019482 (rank : 47) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.018226 (rank : 48) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.017791 (rank : 49) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.017237 (rank : 50) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
PMVK_MOUSE
|
||||||
| NC score | 0.017111 (rank : 51) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1G2, Q8R021, Q9D6K3 | Gene names | Pmvk | |||
|
Domain Architecture |

|
|||||
| Description | Phosphomevalonate kinase (EC 2.7.4.2) (PMKase). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.017062 (rank : 52) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.017024 (rank : 53) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
K1718_HUMAN
|
||||||
| NC score | 0.016418 (rank : 54) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
K1718_MOUSE
|
||||||
| NC score | 0.014504 (rank : 55) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
DIP2A_MOUSE
|
||||||
| NC score | 0.012679 (rank : 56) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
DIP2A_HUMAN
|
||||||
| NC score | 0.012601 (rank : 57) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.012426 (rank : 58) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.011559 (rank : 59) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.011015 (rank : 60) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.011010 (rank : 61) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.010403 (rank : 62) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.010331 (rank : 63) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
HXB1_HUMAN
|
||||||
| NC score | 0.009095 (rank : 64) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P14653 | Gene names | HOXB1, HOX2I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B1 (Hox-2I). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.007601 (rank : 65) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.006072 (rank : 66) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
DLX4_HUMAN
|
||||||
| NC score | 0.004027 (rank : 67) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92988, O60480, Q13265, Q6PJK0, Q9HBE0 | Gene names | DLX4, BP1, DLX7, DLX8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein DLX-4 (DLX-7) (DLX-8) (Beta protein 1). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.003141 (rank : 68) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.003050 (rank : 69) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
RAB6C_HUMAN
|
||||||
| NC score | 0.002161 (rank : 70) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0N0, Q9P128 | Gene names | RAB6C, WTH3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-6C (Rab6-like protein WTH3). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.001038 (rank : 71) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.000981 (rank : 72) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
HGFA_MOUSE
|
||||||
| NC score | 0.000276 (rank : 73) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | -0.001916 (rank : 74) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 74 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||