Please be patient as the page loads
|
NUP53_MOUSE
|
||||||
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NUP53_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
NUP53_HUMAN
|
||||||
| θ value | 2.10606e-174 (rank : 2) | NC score | 0.946720 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 3) | NC score | 0.125060 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 4) | NC score | 0.137782 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.160802 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.137626 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.118924 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.062275 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.051360 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.106567 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.086682 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.086745 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CITE4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.122038 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
DAG1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.121211 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14118 | Gene names | DAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
DAG1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.122482 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
EP400_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.058232 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.027372 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.082076 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
GPR64_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.032566 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.033595 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
DUS8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.033449 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.054017 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.021950 (rank : 105) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.056640 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.046085 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.067709 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.074890 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.054138 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
UBQL2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.047538 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZM0, Q7TSJ8, Q8VDH9 | Gene names | Ubqln2, Plic2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
WASP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.062759 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.078123 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
CO039_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.073364 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.092994 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.024354 (rank : 101) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.033652 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
WASF4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.049198 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.030556 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
EYA4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.036476 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
IL3RB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.054894 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
K0240_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.049902 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.046809 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.038901 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.043093 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.036328 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.053694 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.039632 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MK07_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.008556 (rank : 115) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.027386 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.060723 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.030316 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.029533 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.012043 (rank : 113) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.051251 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.013884 (rank : 111) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.046996 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.051935 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.063820 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.042780 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.051077 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.058228 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.055674 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.051285 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.044960 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.044995 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.056628 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.005198 (rank : 118) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.015161 (rank : 109) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.056525 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.052076 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.018147 (rank : 108) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
VASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.021650 (rank : 106) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.113149 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.022057 (rank : 104) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.018950 (rank : 107) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.012691 (rank : 112) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.045572 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
KLF10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.003082 (rank : 119) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13118 | Gene names | KLF10, TIEG, TIEG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (EGR-alpha). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.045585 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.029956 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.051304 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
APC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.022486 (rank : 103) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.052115 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.028768 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.015082 (rank : 110) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.046905 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.038022 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
LZTS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.011541 (rank : 114) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRK4, Q8N3I0, Q96J79, Q96JL2 | Gene names | LZTS2, KIAA1813, LAPSER1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2 (Protein LAPSER1). | |||||
|
MEF2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.024484 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.006411 (rank : 116) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.036246 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.006061 (rank : 117) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.022513 (rank : 102) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
SSBP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.048835 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |
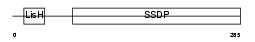
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.048835 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |
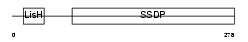
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.050476 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.050915 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.092428 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.056802 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CEL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.051859 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.062533 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
K1802_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.060649 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.065845 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.055339 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.064099 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MISS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.051849 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.089788 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.064448 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.070652 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.064043 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.067161 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.054508 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.075922 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.050818 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.057538 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.081593 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.075600 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050860 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.053881 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.059901 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
NUP53_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
NUP53_HUMAN
|
||||||
| NC score | 0.946720 (rank : 2) | θ value | 2.10606e-174 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NFH5, Q4ZFZ9, Q53S95, Q8TDJ1 | Gene names | NUP35, MP44, NUP53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.160802 (rank : 3) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.137782 (rank : 4) | θ value | 0.000602161 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.137626 (rank : 5) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.125060 (rank : 6) | θ value | 0.00035302 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
DAG1_MOUSE
|
||||||
| NC score | 0.122482 (rank : 7) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
CITE4_MOUSE
|
||||||
| NC score | 0.122038 (rank : 8) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
DAG1_HUMAN
|
||||||
| NC score | 0.121211 (rank : 9) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14118 | Gene names | DAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.118924 (rank : 10) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.113149 (rank : 11) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.106567 (rank : 12) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.092994 (rank : 13) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.092428 (rank : 14) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.089788 (rank : 15) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.086745 (rank : 16) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.086682 (rank : 17) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.082076 (rank : 18) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.081593 (rank : 19) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.078123 (rank : 20) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.075922 (rank : 21) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.075600 (rank : 22) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.074890 (rank : 23) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.073364 (rank : 24) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.070652 (rank : 25) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.067709 (rank : 26) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.067161 (rank : 27) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.065845 (rank : 28) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.064448 (rank : 29) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.064099 (rank : 30) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.064043 (rank : 31) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.063820 (rank : 32) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.062759 (rank : 33) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.062533 (rank : 34) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.062275 (rank : 35) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.060723 (rank : 36) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.060649 (rank : 37) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.059901 (rank : 38) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.058232 (rank : 39) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.058228 (rank : 40) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.057538 (rank : 41) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.056802 (rank : 42) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.056640 (rank : 43) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.056628 (rank : 44) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.056525 (rank : 45) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.055674 (rank : 46) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.055339 (rank : 47) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
IL3RB_HUMAN
|
||||||
| NC score | 0.054894 (rank : 48) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P32927 | Gene names | CSF2RB, IL3RB, IL5RB | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen) (CDw131). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.054508 (rank : 49) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.054138 (rank : 50) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.054017 (rank : 51) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.053881 (rank : 52) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.053694 (rank : 53) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.052115 (rank : 54) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.052076 (rank : 55) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.051935 (rank : 56) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.051859 (rank : 57) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.051849 (rank : 58) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.051360 (rank : 59) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.051304 (rank : 60) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.051285 (rank : 61) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.051251 (rank : 62) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.051077 (rank : 63) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.050915 (rank : 64) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.050860 (rank : 65) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.050818 (rank : 66) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.050476 (rank : 67) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
K0240_MOUSE
|
||||||
| NC score | 0.049902 (rank : 68) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
WASF4_HUMAN
|
||||||
| NC score | 0.049198 (rank : 69) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IV90 | Gene names | WASF4, SCAR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 4 (WASP-family protein member 4). | |||||
|
SSBP3_HUMAN
|
||||||
| NC score | 0.048835 (rank : 70) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| NC score | 0.048835 (rank : 71) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
UBQL2_MOUSE
|
||||||
| NC score | 0.047538 (rank : 72) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZM0, Q7TSJ8, Q8VDH9 | Gene names | Ubqln2, Plic2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.046996 (rank : 73) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.046905 (rank : 74) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.046809 (rank : 75) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.046085 (rank : 76) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.045585 (rank : 77) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.045572 (rank : 78) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.044995 (rank : 79) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.044960 (rank : 80) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.043093 (rank : 81) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.042780 (rank : 82) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.039632 (rank : 83) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.038901 (rank : 84) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.038022 (rank : 85) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
EYA4_MOUSE
|
||||||
| NC score | 0.036476 (rank : 86) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.036328 (rank : 87) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.036246 (rank : 88) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.033652 (rank : 89) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.033595 (rank : 90) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.033449 (rank : 91) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
GPR64_HUMAN
|
||||||
| NC score | 0.032566 (rank : 92) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.030556 (rank : 93) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.030316 (rank : 94) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.029956 (rank : 95) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.029533 (rank : 96) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.028768 (rank : 97) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.027386 (rank : 98) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.027372 (rank : 99) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
MEF2B_HUMAN
|
||||||
| NC score | 0.024484 (rank : 100) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.024354 (rank : 101) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.022513 (rank : 102) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.022486 (rank : 103) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.022057 (rank : 104) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.021950 (rank : 105) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.021650 (rank : 106) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.018950 (rank : 107) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.018147 (rank : 108) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.015161 (rank : 109) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.015082 (rank : 110) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.013884 (rank : 111) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.012691 (rank : 112) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.012043 (rank : 113) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
LZTS2_HUMAN
|
||||||
| NC score | 0.011541 (rank : 114) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRK4, Q8N3I0, Q96J79, Q96JL2 | Gene names | LZTS2, KIAA1813, LAPSER1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2 (Protein LAPSER1). | |||||
|
MK07_HUMAN
|
||||||
| NC score | 0.008556 (rank : 115) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.006411 (rank : 116) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.006061 (rank : 117) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
ZN335_HUMAN
|
||||||
| NC score | 0.005198 (rank : 118) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
KLF10_HUMAN
|
||||||
| NC score | 0.003082 (rank : 119) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13118 | Gene names | KLF10, TIEG, TIEG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (EGR-alpha). | |||||