Please be patient as the page loads
|
GGA3_MOUSE
|
||||||
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GGA3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.975961 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 1.29861e-115 (rank : 3) | NC score | 0.931614 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_MOUSE
|
||||||
| θ value | 3.53647e-113 (rank : 4) | NC score | 0.933405 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 1.65422e-86 (rank : 5) | NC score | 0.920737 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 1.00358e-83 (rank : 6) | NC score | 0.925488 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
TOM1_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 7) | NC score | 0.699684 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 8) | NC score | 0.694402 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 9) | NC score | 0.656398 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TM1L1_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 10) | NC score | 0.646120 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 11) | NC score | 0.368065 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 12) | NC score | 0.374700 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
STAM1_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 13) | NC score | 0.409700 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_MOUSE
|
||||||
| θ value | 2.0648e-12 (rank : 14) | NC score | 0.403938 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM2_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 15) | NC score | 0.416950 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 16) | NC score | 0.412352 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 17) | NC score | 0.360820 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 18) | NC score | 0.411018 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 19) | NC score | 0.353621 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
STAM2_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 20) | NC score | 0.400891 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 21) | NC score | 0.055044 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 22) | NC score | 0.079491 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 23) | NC score | 0.032102 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 24) | NC score | 0.027192 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 25) | NC score | 0.033392 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 26) | NC score | 0.048042 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
CBL_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 27) | NC score | 0.032997 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 28) | NC score | 0.051217 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 29) | NC score | 0.053565 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.042941 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.036838 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.045930 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 33) | NC score | 0.032739 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
K1543_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.056382 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.041545 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | 0.034509 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
DCC_HUMAN
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.023154 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.029029 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.034424 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
AATK_MOUSE
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.008317 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.066787 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CU013_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.050428 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.042322 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.033600 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
PKHG4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.027647 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.035040 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.050783 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.034961 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.027472 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.031082 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.044031 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.034053 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.027444 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.029740 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.033049 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.056508 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.036789 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
MCPH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.028074 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.020985 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.028381 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.035924 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CR034_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.037245 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.033680 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.035128 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SON_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.051344 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.029457 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.037941 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.025049 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.039934 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.039899 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
UBQL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.017351 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.026702 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.039183 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CING_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.025351 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CN092_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.034045 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.029825 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
SOX3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.017061 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
ADAM9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.003939 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13443, Q10718, Q8NFM6 | Gene names | ADAM9, KIAA0021, MCMP, MDC9, MLTNG | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma) (Cellular disintegrin-related protein). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.028038 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.017665 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.013784 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
SUSD4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.018638 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
TEX9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.035060 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
X3CL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.024166 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78423, O00672 | Gene names | CX3CL1, A-152E5.2, FKN, NTT, SCYD1 | |||
|
Domain Architecture |
|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
CTGE2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.030043 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.035814 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.031805 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.024860 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NCOA7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.015025 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NI08, Q3LID6, Q4G0V1, Q5TF95, Q6IPQ4, Q6NE83, Q86T89, Q8N1W4 | Gene names | NCOA7, ERAP140, ESNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7 (140 kDa estrogen receptor-associated protein) (Estrogen nuclear receptor coactivator 1). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.013538 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.024984 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.035681 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.037172 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
AAKG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.010538 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
ANGL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.011561 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5C1 | Gene names | ANGPTL3, ANGPT5 | |||
|
Domain Architecture |
|
|||||
| Description | Angiopoietin-related protein 3 precursor (Angiopoietin-like 3) (Angiopoietin-5) (ANG-5). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.011755 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.031941 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
KLC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.011628 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.015574 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.028775 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.022707 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RB3GP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.014704 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UJ7, Q6A0D7, Q8C4Y0, Q8C679, Q8C6A5, Q8K324 | Gene names | Rab3gap1, Kiaa0066, Rab3gap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein catalytic subunit (RAB3 GTPase- activating protein 130 kDa subunit) (Rab3-GAP p130) (Rab3-GAP). | |||||
|
TBCD4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.012472 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BYJ6, Q5DU23, Q66JU2, Q6P2M2, Q8BMH6, Q8BXM2 | Gene names | Tbc1d4, Kiaa0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.020614 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CI039_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.022848 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CKAP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.025177 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.032006 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
K0226_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.013019 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.023831 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.019192 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.048953 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.033528 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.028919 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.004362 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
STMN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.016698 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
STMN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.016521 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O70166 | Gene names | Stmn3, Sclip | |||
|
Domain Architecture |
|
|||||
| Description | Stathmin-3 (SCG10-like protein) (SCG10-related protein HiAT3) (Hippocampus abundant transcript 3). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.031926 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TMM24_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.020263 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.019532 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
WASL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.048530 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.046740 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.020198 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CCD93_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.025742 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.025314 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.053514 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.024424 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.025785 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
LAMA5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.005819 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.023509 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.024989 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NUP53_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.012691 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
NUP98_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.014078 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.021817 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
SAPS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.009268 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R3Q2, Q9CXB7 | Gene names | Saps2, Kiaa0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
BC11B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | -0.001781 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PV8, Q8C2I1, Q99PV6, Q99PV7, Q9JLF8 | Gene names | Bcl11b, Ctip2, Rit1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (mRit1) (COUP-TF-interacting protein 2). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.007275 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.017790 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.011507 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.011293 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.034927 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.008336 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.023093 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.023355 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.023421 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.023110 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
NOS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.005400 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29474, Q13662, Q14251, Q14434 | Gene names | NOS3 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
PLK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | -0.003709 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60806, Q60822, Q9R009 | Gene names | Plk3, Cnk, Fnk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase). | |||||
|
ZSCA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | -0.003897 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |
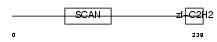
|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||
|
ALEX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.050275 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
AP2A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.104588 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.102301 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.100066 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.099963 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.092308 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.093953 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP4E1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.103845 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.102867 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.073575 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.071114 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.050024 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.054885 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.053381 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.975961 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.933405 (rank : 3) | θ value | 3.53647e-113 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.931614 (rank : 4) | θ value | 1.29861e-115 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.925488 (rank : 5) | θ value | 1.00358e-83 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.920737 (rank : 6) | θ value | 1.65422e-86 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
TOM1_MOUSE
|
||||||
| NC score | 0.699684 (rank : 7) | θ value | 7.56453e-23 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| NC score | 0.694402 (rank : 8) | θ value | 1.09232e-21 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.656398 (rank : 9) | θ value | 3.40345e-15 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TM1L1_HUMAN
|
||||||
| NC score | 0.646120 (rank : 10) | θ value | 1.29331e-14 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
STAM2_HUMAN
|
||||||
| NC score | 0.416950 (rank : 11) | θ value | 2.0648e-12 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.412352 (rank : 12) | θ value | 4.59992e-12 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.411018 (rank : 13) | θ value | 7.84624e-12 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
STAM1_HUMAN
|
||||||
| NC score | 0.409700 (rank : 14) | θ value | 1.58096e-12 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_MOUSE
|
||||||
| NC score | 0.403938 (rank : 15) | θ value | 2.0648e-12 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM2_MOUSE
|
||||||
| NC score | 0.400891 (rank : 16) | θ value | 6.64225e-11 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.374700 (rank : 17) | θ value | 4.91457e-14 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.368065 (rank : 18) | θ value | 3.76295e-14 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.360820 (rank : 19) | θ value | 6.00763e-12 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.353621 (rank : 20) | θ value | 1.33837e-11 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.104588 (rank : 21) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP4E1_HUMAN
|
||||||
| NC score | 0.103845 (rank : 22) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| NC score | 0.102867 (rank : 23) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| NC score | 0.102301 (rank : 24) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.100066 (rank : 25) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| NC score | 0.099963 (rank : 26) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.093953 (rank : 27) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.092308 (rank : 28) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.079491 (rank : 29) | θ value | 0.00390308 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.073575 (rank : 30) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.071114 (rank : 31) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.066787 (rank : 32) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056508 (rank : 33) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.056382 (rank : 34) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.055044 (rank : 35) | θ value | 0.00134147 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.054885 (rank : 36) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.053565 (rank : 37) | θ value | 0.0330416 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.053514 (rank : 38) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.053381 (rank : 39) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.051344 (rank : 40) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.051217 (rank : 41) | θ value | 0.0330416 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.050783 (rank : 42) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CU013_HUMAN
|
||||||
| NC score | 0.050428 (rank : 43) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
ALEX_MOUSE
|
||||||
| NC score | 0.050275 (rank : 44) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6R0H6, Q8BIR3 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.050024 (rank : 45) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.048953 (rank : 46) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.048530 (rank : 47) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.048042 (rank : 48) | θ value | 0.0252991 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.046740 (rank : 49) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.045930 (rank : 50) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.044031 (rank : 51) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.042941 (rank : 52) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.042322 (rank : 53) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.041545 (rank : 54) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.039934 (rank : 55) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.039899 (rank : 56) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.039183 (rank : 57) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.037941 (rank : 58) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CR034_MOUSE
|
||||||
| NC score | 0.037245 (rank : 59) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.037172 (rank : 60) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.036838 (rank : 61) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.036789 (rank : 62) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.035924 (rank : 63) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.035814 (rank : 64) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.035681 (rank : 65) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.035128 (rank : 66) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TEX9_HUMAN
|
||||||
| NC score | 0.035060 (rank : 67) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.035040 (rank : 68) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.034961 (rank : 69) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.034927 (rank : 70) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.034509 (rank : 71) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.034424 (rank : 72) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.034053 (rank : 73) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.034045 (rank : 74) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.033680 (rank : 75) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.033600 (rank : 76) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.033528 (rank : 77) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.033392 (rank : 78) | θ value | 0.0113563 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.033049 (rank : 79) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.032997 (rank : 80) | θ value | 0.0252991 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.032739 (rank : 81) | θ value | 0.0961366 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.032102 (rank : 82) | θ value | 0.00390308 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.032006 (rank : 83) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.031941 (rank : 84) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.031926 (rank : 85) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.031805 (rank : 86) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.031082 (rank : 87) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
CTGE2_HUMAN
|
||||||
| NC score | 0.030043 (rank : 88) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.029825 (rank : 89) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.029740 (rank : 90) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.029457 (rank : 91) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.029029 (rank : 92) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.028919 (rank : 93) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.028775 (rank : 94) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.028381 (rank : 95) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
MCPH1_HUMAN
|
||||||
| NC score | 0.028074 (rank : 96) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.028038 (rank : 97) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
PKHG4_HUMAN
|
||||||
| NC score | 0.027647 (rank : 98) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.027472 (rank : 99) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.027444 (rank : 100) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.027192 (rank : 101) | θ value | 0.0113563 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.026702 (rank : 102) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.025785 (rank : 103) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CCD93_HUMAN
|
||||||
| NC score | 0.025742 (rank : 104) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.025351 (rank : 105) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.025314 (rank : 106) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
CKAP2_MOUSE
|
||||||
| NC score | 0.025177 (rank : 107) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.025049 (rank : 108) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.024989 (rank : 109) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.024984 (rank : 110) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.024860 (rank : 111) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.024424 (rank : 112) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
X3CL1_HUMAN
|
||||||
| NC score | 0.024166 (rank : 113) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78423, O00672 | Gene names | CX3CL1, A-152E5.2, FKN, NTT, SCYD1 | |||
|
Domain Architecture |

|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.023831 (rank : 114) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.023509 (rank : 115) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.023421 (rank : 116) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.023355 (rank : 117) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
DCC_HUMAN
|
||||||
| NC score | 0.023154 (rank : 118) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.023110 (rank : 119) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.023093 (rank : 120) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.022848 (rank : 121) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.022707 (rank : 122) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.021817 (rank : 123) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.020985 (rank : 124) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.020614 (rank : 125) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
TMM24_MOUSE
|
||||||
| NC score | 0.020263 (rank : 126) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80X80 | Gene names | Tmem24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 24. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.020198 (rank : 127) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.019532 (rank : 128) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.019192 (rank : 129) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
SUSD4_MOUSE
|
||||||
| NC score | 0.018638 (rank : 130) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.017790 (rank : 131) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.017665 (rank : 132) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
UBQL3_HUMAN
|
||||||
| NC score | 0.017351 (rank : 133) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
SOX3_HUMAN
|
||||||
| NC score | 0.017061 (rank : 134) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
STMN3_HUMAN
|
||||||
| NC score | 0.016698 (rank : 135) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZ72, O75527, Q969Y4 | Gene names | STMN3, SCLIP | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-3 (SCG10-like protein). | |||||
|
STMN3_MOUSE
|
||||||
| NC score | 0.016521 (rank : 136) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O70166 | Gene names | Stmn3, Sclip | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-3 (SCG10-like protein) (SCG10-related protein HiAT3) (Hippocampus abundant transcript 3). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.015574 (rank : 137) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
NCOA7_HUMAN
|
||||||
| NC score | 0.015025 (rank : 138) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NI08, Q3LID6, Q4G0V1, Q5TF95, Q6IPQ4, Q6NE83, Q86T89, Q8N1W4 | Gene names | NCOA7, ERAP140, ESNA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7 (140 kDa estrogen receptor-associated protein) (Estrogen nuclear receptor coactivator 1). | |||||
|
RB3GP_MOUSE
|
||||||
| NC score | 0.014704 (rank : 139) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UJ7, Q6A0D7, Q8C4Y0, Q8C679, Q8C6A5, Q8K324 | Gene names | Rab3gap1, Kiaa0066, Rab3gap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein catalytic subunit (RAB3 GTPase- activating protein 130 kDa subunit) (Rab3-GAP p130) (Rab3-GAP). | |||||
|
NUP98_HUMAN
|
||||||
| NC score | 0.014078 (rank : 140) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.013784 (rank : 141) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.013538 (rank : 142) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
K0226_HUMAN
|
||||||
| NC score | 0.013019 (rank : 143) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
NUP53_MOUSE
|
||||||
| NC score | 0.012691 (rank : 144) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R4R6, Q9D7J2 | Gene names | Nup35, Mp44, Nup53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NUP53 (Nuclear pore complex protein Nup53) (Nucleoporin Nup35) (35 kDa nucleoporin) (Mitotic phosphoprotein 44) (MP-44). | |||||
|
TBCD4_MOUSE
|
||||||
| NC score | 0.012472 (rank : 145) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BYJ6, Q5DU23, Q66JU2, Q6P2M2, Q8BMH6, Q8BXM2 | Gene names | Tbc1d4, Kiaa0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.011755 (rank : 146) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.011628 (rank : 147) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
ANGL3_HUMAN
|
||||||
| NC score | 0.011561 (rank : 148) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5C1 | Gene names | ANGPTL3, ANGPT5 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 3 precursor (Angiopoietin-like 3) (Angiopoietin-5) (ANG-5). | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.011507 (rank : 149) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.011293 (rank : 150) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
AAKG2_HUMAN
|
||||||
| NC score | 0.010538 (rank : 151) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
SAPS2_MOUSE
|
||||||
| NC score | 0.009268 (rank : 152) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R3Q2, Q9CXB7 | Gene names | Saps2, Kiaa0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.008336 (rank : 153) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
AATK_MOUSE
|
||||||
| NC score | 0.008317 (rank : 154) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.007275 (rank : 155) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
LAMA5_HUMAN
|
||||||
| NC score | 0.005819 (rank : 156) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
NOS3_HUMAN
|
||||||
| NC score | 0.005400 (rank : 157) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29474, Q13662, Q14251, Q14434 | Gene names | NOS3 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.004362 (rank : 158) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
ADAM9_HUMAN
|
||||||
| NC score | 0.003939 (rank : 159) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13443, Q10718, Q8NFM6 | Gene names | ADAM9, KIAA0021, MCMP, MDC9, MLTNG | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma) (Cellular disintegrin-related protein). | |||||
|
BC11B_MOUSE
|
||||||
| NC score | -0.001781 (rank : 160) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PV8, Q8C2I1, Q99PV6, Q99PV7, Q9JLF8 | Gene names | Bcl11b, Ctip2, Rit1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (mRit1) (COUP-TF-interacting protein 2). | |||||
|
PLK3_MOUSE
|
||||||
| NC score | -0.003709 (rank : 161) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60806, Q60822, Q9R009 | Gene names | Plk3, Cnk, Fnk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase). | |||||
|
ZSCA2_MOUSE
|
||||||
| NC score | -0.003897 (rank : 162) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 767 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07230 | Gene names | Zscan2, Zfp-29, Zfp29 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and SCAN domain-containing protein 2 (Zinc finger protein 29) (Zfp-29). | |||||