Please be patient as the page loads
|
TOM1_MOUSE
|
||||||
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TOM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991669 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_HUMAN
|
||||||
| θ value | 3.71096e-62 (rank : 3) | NC score | 0.913547 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 2.03627e-60 (rank : 4) | NC score | 0.910587 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 5) | NC score | 0.720624 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA1_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 6) | NC score | 0.690628 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 7) | NC score | 0.699684 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
STAM1_HUMAN
|
||||||
| θ value | 2.20094e-22 (rank : 8) | NC score | 0.532609 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 9) | NC score | 0.525793 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 10) | NC score | 0.687548 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
STAM2_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 11) | NC score | 0.541267 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
STAM2_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 12) | NC score | 0.526130 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 13) | NC score | 0.485882 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 14) | NC score | 0.483619 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 15) | NC score | 0.666493 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 16) | NC score | 0.662699 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.031773 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.015912 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.031678 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.021075 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.032017 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ESAM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.018916 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
SSNA1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.053894 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJ94 | Gene names | Ssna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sjoegren syndrome nuclear autoantigen 1 homolog. | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.031005 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.024419 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.029863 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.014711 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
RLA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.045606 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05386 | Gene names | RPLP1, RRP1 | |||
|
Domain Architecture |

|
|||||
| Description | 60S acidic ribosomal protein P1. | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.030848 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
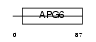
SSNA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.044605 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43805, Q6FG70, Q9BVW8 | Gene names | SSNA1, NA14 | |||
|
Domain Architecture |
|
|||||
| Description | Sjoegren syndrome nuclear autoantigen 1 (Nuclear autoantigen of 14 kDa). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.025521 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PCBP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.014736 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
STRC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.017577 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.007461 (rank : 63) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.005358 (rank : 67) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
GOG8A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.016967 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
LRCH3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.006204 (rank : 65) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96II8, Q96FP9, Q9NT52 | Gene names | LRCH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
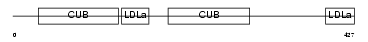
LRP10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.008912 (rank : 61) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQH7, Q7TS95, Q921T0, Q9EPE8 | Gene names | Lrp10, Lrp9 | |||
|
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 10 precursor. | |||||
|
RGL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.011231 (rank : 59) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61193, Q9QUJ2 | Gene names | Rgl2, Rab2l, Rlf | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.015977 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.015929 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
CENPK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.014598 (rank : 57) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BS16, Q9H4L0 | Gene names | CENPK, ICEN37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (Interphase centromere complex protein 37) (Protein AF-5alpha) (p33). | |||||
|
GP1BA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.005016 (rank : 68) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
RLA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.037791 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47955 | Gene names | Rplp1 | |||
|
Domain Architecture |

|
|||||
| Description | 60S acidic ribosomal protein P1. | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.007016 (rank : 64) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
PCBP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.012504 (rank : 58) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
U171_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.018762 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12980, Q4TT56, Q92469 | Gene names | CGTHBA, C16orf35 | |||
|
Domain Architecture |

|
|||||
| Description | CGTHBA protein (-14 gene protein). | |||||
|
VDP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.018788 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
CHM4B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.009773 (rank : 60) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D8B3, Q3TXM7, Q91VM7, Q922P1 | Gene names | Chmp4b | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.004661 (rank : 69) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
DYSF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.005435 (rank : 66) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
LZTS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.007957 (rank : 62) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
ZFP91_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | -0.002118 (rank : 70) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
ABI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.051258 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CBW3, Q60747, Q91ZM5, Q923I9, Q99KH4 | Gene names | Abi1, Ssh3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Ablphilin-1). | |||||
|
ABI3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.050083 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.102626 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.098897 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.088499 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.093879 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
DBNL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.058651 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJU6, P84070, Q6IAI8, Q96F30, Q96K74, Q9HBN8, Q9NR72 | Gene names | DBNL, CMAP, SH3P7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Drebrin F) (Cervical SH3P7) (HPK1-interacting protein of 55 kDa) (HIP-55) (Cervical mucin-associated protein). | |||||
|
DBNL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.055329 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
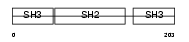
GRAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.057735 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75791, O43726 | Gene names | GRAP2, GADS, GRB2L, GRID | |||
|
Domain Architecture |
|
|||||
| Description | GRB2-related adapter protein 2 (GADS protein) (Growth factor receptor- binding protein) (GRBLG) (Grf40 adapter protein) (Grf-40) (GRB-2-like protein) (GRB2L) (GRBX) (P38) (Hematopoietic cell-associated adapter protein GrpL) (Adapter protein GRID) (SH3-SH2-SH3 adapter Mona). | |||||
|
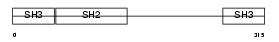
GRAP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.057603 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89100 | Gene names | Grap2, Gads, Grb2l, Grid, Mona | |||
|
Domain Architecture |
|
|||||
| Description | GRB2-related adaptor protein 2 (GADS protein) (Growth factor receptor- binding protein) (GRBLG) (GRB-2-like protein) (GRB2L) (Hematopoietic cell-associated adaptor protein GrpL) (GRB-2-related monocytic adapter protein) (Monocytic adapter) (MONA) (Adapter protein GRID). | |||||
|
NCF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.059043 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19878, Q2PP06, Q8NFC7, Q9BV51 | Gene names | NCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 2 (NCF-2) (Neutrophil NADPH oxidase factor 2) (67 kDa neutrophil oxidase factor) (p67-phox) (NOXA2). | |||||
|
NCF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.053545 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97369 | Gene names | Ncf4 | |||
|
Domain Architecture |
|
|||||
| Description | Neutrophil cytosol factor 4 (NCF-4) (Neutrophil NADPH oxidase factor 4) (p40-phox) (p40phox). | |||||
|
SRC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.052694 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14247 | Gene names | CTTN, EMS1 | |||
|
Domain Architecture |
|
|||||
| Description | Src substrate cortactin (Amplaxin) (Oncogene EMS1). | |||||
|
STAC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.064022 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96MF2, Q96HU5 | Gene names | STAC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
STAC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.064994 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ71 | Gene names | Stac3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
STAC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.054725 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99469 | Gene names | STAC | |||
|
Domain Architecture |
|
|||||
| Description | SH3 and cysteine-rich domain-containing protein (SRC homology 3 and cysteine-rich domain protein). | |||||
|
STAC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.054514 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97306 | Gene names | Stac | |||
|
Domain Architecture |
|
|||||
| Description | SH3 and cysteine-rich domain-containing protein (SRC homology 3 and cysteine-rich domain protein). | |||||
|
TOM1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| NC score | 0.991669 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_HUMAN
|
||||||
| NC score | 0.913547 (rank : 3) | θ value | 3.71096e-62 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.910587 (rank : 4) | θ value | 2.03627e-60 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.720624 (rank : 5) | θ value | 1.24688e-25 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.699684 (rank : 6) | θ value | 7.56453e-23 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.690628 (rank : 7) | θ value | 7.56453e-23 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.687548 (rank : 8) | θ value | 7.0802e-21 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.666493 (rank : 9) | θ value | 3.07829e-16 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.662699 (rank : 10) | θ value | 1.68911e-14 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
STAM2_HUMAN
|
||||||
| NC score | 0.541267 (rank : 11) | θ value | 2.69047e-20 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
STAM1_HUMAN
|
||||||
| NC score | 0.532609 (rank : 12) | θ value | 2.20094e-22 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM2_MOUSE
|
||||||
| NC score | 0.526130 (rank : 13) | θ value | 2.69047e-20 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
STAM1_MOUSE
|
||||||
| NC score | 0.525793 (rank : 14) | θ value | 2.87452e-22 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.485882 (rank : 15) | θ value | 8.10077e-17 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.483619 (rank : 16) | θ value | 8.10077e-17 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.102626 (rank : 17) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.098897 (rank : 18) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.093879 (rank : 19) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.088499 (rank : 20) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
STAC3_MOUSE
|
||||||
| NC score | 0.064994 (rank : 21) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ71 | Gene names | Stac3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
STAC3_HUMAN
|
||||||
| NC score | 0.064022 (rank : 22) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96MF2, Q96HU5 | Gene names | STAC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
NCF2_HUMAN
|
||||||
| NC score | 0.059043 (rank : 23) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19878, Q2PP06, Q8NFC7, Q9BV51 | Gene names | NCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 2 (NCF-2) (Neutrophil NADPH oxidase factor 2) (67 kDa neutrophil oxidase factor) (p67-phox) (NOXA2). | |||||
|
DBNL_HUMAN
|
||||||
| NC score | 0.058651 (rank : 24) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJU6, P84070, Q6IAI8, Q96F30, Q96K74, Q9HBN8, Q9NR72 | Gene names | DBNL, CMAP, SH3P7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Drebrin F) (Cervical SH3P7) (HPK1-interacting protein of 55 kDa) (HIP-55) (Cervical mucin-associated protein). | |||||
|
GRAP2_HUMAN
|
||||||
| NC score | 0.057735 (rank : 25) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75791, O43726 | Gene names | GRAP2, GADS, GRB2L, GRID | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-related adapter protein 2 (GADS protein) (Growth factor receptor- binding protein) (GRBLG) (Grf40 adapter protein) (Grf-40) (GRB-2-like protein) (GRB2L) (GRBX) (P38) (Hematopoietic cell-associated adapter protein GrpL) (Adapter protein GRID) (SH3-SH2-SH3 adapter Mona). | |||||
|
GRAP2_MOUSE
|
||||||
| NC score | 0.057603 (rank : 26) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89100 | Gene names | Grap2, Gads, Grb2l, Grid, Mona | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-related adaptor protein 2 (GADS protein) (Growth factor receptor- binding protein) (GRBLG) (GRB-2-like protein) (GRB2L) (Hematopoietic cell-associated adaptor protein GrpL) (GRB-2-related monocytic adapter protein) (Monocytic adapter) (MONA) (Adapter protein GRID). | |||||
|
DBNL_MOUSE
|
||||||
| NC score | 0.055329 (rank : 27) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
STAC_HUMAN
|
||||||
| NC score | 0.054725 (rank : 28) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99469 | Gene names | STAC | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and cysteine-rich domain-containing protein (SRC homology 3 and cysteine-rich domain protein). | |||||
|
STAC_MOUSE
|
||||||
| NC score | 0.054514 (rank : 29) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97306 | Gene names | Stac | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and cysteine-rich domain-containing protein (SRC homology 3 and cysteine-rich domain protein). | |||||
|
SSNA1_MOUSE
|
||||||
| NC score | 0.053894 (rank : 30) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJ94 | Gene names | Ssna1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sjoegren syndrome nuclear autoantigen 1 homolog. | |||||
|
NCF4_MOUSE
|
||||||
| NC score | 0.053545 (rank : 31) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97369 | Gene names | Ncf4 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 4 (NCF-4) (Neutrophil NADPH oxidase factor 4) (p40-phox) (p40phox). | |||||
|
SRC8_HUMAN
|
||||||
| NC score | 0.052694 (rank : 32) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14247 | Gene names | CTTN, EMS1 | |||
|
Domain Architecture |
|
|||||
| Description | Src substrate cortactin (Amplaxin) (Oncogene EMS1). | |||||
|
ABI1_MOUSE
|
||||||
| NC score | 0.051258 (rank : 33) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CBW3, Q60747, Q91ZM5, Q923I9, Q99KH4 | Gene names | Abi1, Ssh3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Ablphilin-1). | |||||
|
ABI3_MOUSE
|
||||||
| NC score | 0.050083 (rank : 34) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYZ1, Q6PE63, Q9D7S4 | Gene names | Abi3, Nesh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
RLA1_HUMAN
|
||||||
| NC score | 0.045606 (rank : 35) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05386 | Gene names | RPLP1, RRP1 | |||
|
Domain Architecture |

|
|||||
| Description | 60S acidic ribosomal protein P1. | |||||
|
SSNA1_HUMAN
|
||||||
| NC score | 0.044605 (rank : 36) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43805, Q6FG70, Q9BVW8 | Gene names | SSNA1, NA14 | |||
|
Domain Architecture |

|
|||||
| Description | Sjoegren syndrome nuclear autoantigen 1 (Nuclear autoantigen of 14 kDa). | |||||
|
RLA1_MOUSE
|
||||||
| NC score | 0.037791 (rank : 37) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47955 | Gene names | Rplp1 | |||
|
Domain Architecture |

|
|||||
| Description | 60S acidic ribosomal protein P1. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.032017 (rank : 38) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.031773 (rank : 39) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.031678 (rank : 40) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.031005 (rank : 41) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.030848 (rank : 42) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.029863 (rank : 43) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.025521 (rank : 44) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.024419 (rank : 45) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.021075 (rank : 46) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
ESAM_HUMAN
|
||||||
| NC score | 0.018916 (rank : 47) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
VDP_HUMAN
|
||||||
| NC score | 0.018788 (rank : 48) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
U171_HUMAN
|
||||||
| NC score | 0.018762 (rank : 49) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12980, Q4TT56, Q92469 | Gene names | CGTHBA, C16orf35 | |||
|
Domain Architecture |

|
|||||
| Description | CGTHBA protein (-14 gene protein). | |||||
|
STRC_HUMAN
|
||||||
| NC score | 0.017577 (rank : 50) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
GOG8A_HUMAN
|
||||||
| NC score | 0.016967 (rank : 51) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.015977 (rank : 52) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.015929 (rank : 53) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.015912 (rank : 54) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
PCBP4_HUMAN
|
||||||
| NC score | 0.014736 (rank : 55) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57723, Q96AH7 | Gene names | PCBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.014711 (rank : 56) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
CENPK_HUMAN
|
||||||
| NC score | 0.014598 (rank : 57) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BS16, Q9H4L0 | Gene names | CENPK, ICEN37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (Interphase centromere complex protein 37) (Protein AF-5alpha) (p33). | |||||
|
PCBP4_MOUSE
|
||||||
| NC score | 0.012504 (rank : 58) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57724, Q91VR3 | Gene names | Pcbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Poly(rC)-binding protein 4 (Alpha-CP4). | |||||
|
RGL2_MOUSE
|
||||||
| NC score | 0.011231 (rank : 59) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61193, Q9QUJ2 | Gene names | Rgl2, Rab2l, Rlf | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 2 (RalGDS-like factor) (RAS-associated protein RAB2L). | |||||
|
CHM4B_MOUSE
|
||||||
| NC score | 0.009773 (rank : 60) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D8B3, Q3TXM7, Q91VM7, Q922P1 | Gene names | Chmp4b | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4b (Chromatin-modifying protein 4b) (CHMP4b). | |||||
|
LRP10_MOUSE
|
||||||
| NC score | 0.008912 (rank : 61) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQH7, Q7TS95, Q921T0, Q9EPE8 | Gene names | Lrp10, Lrp9 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 10 precursor. | |||||
|
LZTS1_HUMAN
|
||||||
| NC score | 0.007957 (rank : 62) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.007461 (rank : 63) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.007016 (rank : 64) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
LRCH3_HUMAN
|
||||||
| NC score | 0.006204 (rank : 65) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96II8, Q96FP9, Q9NT52 | Gene names | LRCH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
DYSF_MOUSE
|
||||||
| NC score | 0.005435 (rank : 66) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.005358 (rank : 67) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
GP1BA_HUMAN
|
||||||
| NC score | 0.005016 (rank : 68) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.004661 (rank : 69) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
ZFP91_HUMAN
|
||||||
| NC score | -0.002118 (rank : 70) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||