Please be patient as the page loads
|
GGA1_MOUSE
|
||||||
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GGA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.975245 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 1.38103e-133 (rank : 3) | NC score | 0.948066 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 3.4095e-124 (rank : 4) | NC score | 0.952732 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 2.07327e-113 (rank : 5) | NC score | 0.943132 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 3.53647e-113 (rank : 6) | NC score | 0.933405 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
TOM1_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 7) | NC score | 0.690628 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 8) | NC score | 0.685166 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 9) | NC score | 0.654495 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 10) | NC score | 0.362267 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
TM1L1_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 11) | NC score | 0.642276 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 12) | NC score | 0.352910 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 13) | NC score | 0.401718 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 14) | NC score | 0.402093 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
STAM1_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 15) | NC score | 0.379485 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 16) | NC score | 0.383695 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 17) | NC score | 0.342367 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
STAM2_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 18) | NC score | 0.389777 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
STAM2_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 19) | NC score | 0.374890 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 20) | NC score | 0.333381 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 21) | NC score | 0.082230 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 22) | NC score | 0.041708 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 23) | NC score | 0.082722 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 24) | NC score | 0.069421 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 25) | NC score | 0.046062 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 26) | NC score | 0.065195 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 27) | NC score | 0.044931 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.035613 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.035353 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.039832 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
KRHB5_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.020054 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78386, Q9NSB1 | Gene names | KRTHB5 | |||
|
Domain Architecture |
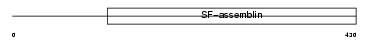
|
|||||
| Description | Keratin, type II cuticular Hb5 (Hair keratin, type II Hb5). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.053075 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
IL4RA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.050662 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |
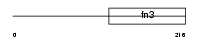
|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.021137 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.040055 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.023216 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.043971 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.059119 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.027174 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
AZI1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.036926 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.033316 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.000415 (rank : 136) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.033116 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.041421 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.026578 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ANGL5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.019654 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XS5, Q86VR9 | Gene names | ANGPTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 5 precursor (Angiopoietin-like 5). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.028873 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.021107 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
IPR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.025037 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
KINH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.033670 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.031564 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.023899 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.022896 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.035576 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.043303 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.027451 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.020704 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.041231 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ANTR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.031031 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |
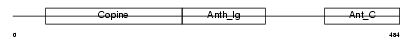
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
ANTR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.032804 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |
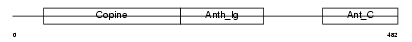
|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.032700 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.039054 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.031710 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
FAK2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.004027 (rank : 131) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
FOXP4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.011771 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
LMBL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.013703 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59178 | Gene names | L3mbtl2 | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like 2 protein (L(3)mbt-like 2 protein) (H-l(3)mbt-like protein). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.028074 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.017478 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.041542 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.016530 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
TLK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.009796 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
TLK2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.009696 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55047, Q9D5Y5 | Gene names | Tlk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2). | |||||
|
KRHB3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.015914 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.022270 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.033494 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SH3G2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.031908 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62420 | Gene names | Sh3gl2, Sh3d2a | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 2 (EC 2.3.1.-) (Endophilin-1) (Endophilin-A1) (SH3 domain protein 2A) (SH3p4). | |||||
|
CN092_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.019418 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.027148 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
FAK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.003750 (rank : 133) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.009499 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.027272 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.005177 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.026676 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.030890 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TLK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.008777 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKI8, Q14150, Q8N591, Q9NYH2, Q9Y4F6 | Gene names | TLK1, KIAA0137 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1) (PKU-beta). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.033033 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DAAM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.014238 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.025207 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
K1C10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.017881 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.032568 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.003361 (rank : 134) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
SON_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.050021 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | -0.000853 (rank : 137) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
AB1IP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.015543 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.023339 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
FOXP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.013855 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.012827 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.011279 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.025246 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.021036 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.028429 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.005212 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
EXOC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.023011 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NV70, Q8WUE7, Q96T15, Q9NZE4 | Gene names | EXOC1, SEC3, SEC3L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.015272 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.023538 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MITF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.008301 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.016023 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PTPRS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.000595 (rank : 135) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
RMD5B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.011647 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YQ7 | Gene names | Rmnd5b | |||
|
Domain Architecture |

|
|||||
| Description | RMD5 homolog B. | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.021171 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
TNR25_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.010549 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.033903 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CAN7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.005793 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
CN092_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.032152 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.025670 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KRHB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.014855 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
KRHB6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.014851 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.021626 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.021448 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NICN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.010895 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSH3, Q8IZQ2 | Gene names | NICN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicolin-1 (Tubulin polyglutamylase complex subunit 5) (PGs5) (NPCEDRG). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.019921 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SEPT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.004606 (rank : 130) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15019, Q14132, Q8IUK9, Q96CB0 | Gene names | SEPT2, DIFF6, KIAA0158, NEDD5 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-2 (Protein NEDD5). | |||||
|
SEPT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.004612 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42208, Q3U9Y5 | Gene names | Sept2, Nedd-5, Nedd5 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-2 (Protein NEDD5) (Neural precursor cell expressed developmentally down-regulated protein 5). | |||||
|
SSH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.003892 (rank : 132) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.016545 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.007484 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TLK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.008027 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C0V0 | Gene names | Tlk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.031643 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
AP2A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.090237 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.089195 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.086745 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.087034 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.077668 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.081639 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP4E1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.090660 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.090017 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.051733 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.975245 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.952732 (rank : 3) | θ value | 3.4095e-124 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.948066 (rank : 4) | θ value | 1.38103e-133 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.943132 (rank : 5) | θ value | 2.07327e-113 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.933405 (rank : 6) | θ value | 3.53647e-113 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
TOM1_MOUSE
|
||||||
| NC score | 0.690628 (rank : 7) | θ value | 7.56453e-23 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| NC score | 0.685166 (rank : 8) | θ value | 1.42661e-21 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.654495 (rank : 9) | θ value | 6.85773e-16 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TM1L1_HUMAN
|
||||||
| NC score | 0.642276 (rank : 10) | θ value | 1.42992e-13 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.402093 (rank : 11) | θ value | 8.67504e-11 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.401718 (rank : 12) | θ value | 5.08577e-11 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
STAM2_HUMAN
|
||||||
| NC score | 0.389777 (rank : 13) | θ value | 2.36244e-08 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
STAM1_HUMAN
|
||||||
| NC score | 0.383695 (rank : 14) | θ value | 1.63604e-09 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_MOUSE
|
||||||
| NC score | 0.379485 (rank : 15) | θ value | 9.59137e-10 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM2_MOUSE
|
||||||
| NC score | 0.374890 (rank : 16) | θ value | 3.08544e-08 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.362267 (rank : 17) | θ value | 2.20605e-14 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.352910 (rank : 18) | θ value | 2.28291e-11 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.342367 (rank : 19) | θ value | 1.38499e-08 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.333381 (rank : 20) | θ value | 6.87365e-08 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP4E1_HUMAN
|
||||||
| NC score | 0.090660 (rank : 21) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.090237 (rank : 22) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| NC score | 0.090017 (rank : 23) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| NC score | 0.089195 (rank : 24) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A2_MOUSE
|
||||||
| NC score | 0.087034 (rank : 25) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.086745 (rank : 26) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.082722 (rank : 27) | θ value | 0.00298849 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.082230 (rank : 28) | θ value | 0.00102713 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.081639 (rank : 29) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.077668 (rank : 30) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.069421 (rank : 31) | θ value | 0.00665767 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.065195 (rank : 32) | θ value | 0.0330416 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.059119 (rank : 33) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.053075 (rank : 34) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.051733 (rank : 35) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
IL4RA_HUMAN
|
||||||
| NC score | 0.050662 (rank : 36) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.050021 (rank : 37) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.046062 (rank : 38) | θ value | 0.0113563 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.044931 (rank : 39) | θ value | 0.0330416 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.043971 (rank : 40) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.043303 (rank : 41) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.041708 (rank : 42) | θ value | 0.00134147 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.041542 (rank : 43) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.041421 (rank : 44) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.041231 (rank : 45) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.040055 (rank : 46) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.039832 (rank : 47) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.039054 (rank : 48) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.036926 (rank : 49) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.035613 (rank : 50) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.035576 (rank : 51) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.035353 (rank : 52) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.033903 (rank : 53) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.033670 (rank : 54) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.033494 (rank : 55) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.033316 (rank : 56) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.033116 (rank : 57) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.033033 (rank : 58) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ANTR1_MOUSE
|
||||||
| NC score | 0.032804 (rank : 59) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CZ52 | Gene names | Antxr1, Atr, Tem8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.032700 (rank : 60) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.032568 (rank : 61) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.032152 (rank : 62) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
SH3G2_MOUSE
|
||||||
| NC score | 0.031908 (rank : 63) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62420 | Gene names | Sh3gl2, Sh3d2a | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 2 (EC 2.3.1.-) (Endophilin-1) (Endophilin-A1) (SH3 domain protein 2A) (SH3p4). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.031710 (rank : 64) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.031643 (rank : 65) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.031564 (rank : 66) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
ANTR1_HUMAN
|
||||||
| NC score | 0.031031 (rank : 67) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H6X2, Q96P02, Q9NVP3 | Gene names | ANTXR1, ATR, TEM8 | |||
|
Domain Architecture |

|
|||||
| Description | Anthrax toxin receptor 1 precursor (Tumor endothelial marker 8). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.030890 (rank : 68) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.028873 (rank : 69) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.028429 (rank : 70) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.028074 (rank : 71) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.027451 (rank : 72) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.027272 (rank : 73) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.027174 (rank : 74) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.027148 (rank : 75) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.026676 (rank : 76) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.026578 (rank : 77) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.025670 (rank : 78) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.025246 (rank : 79) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.025207 (rank : 80) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
IPR1_MOUSE
|
||||||
| NC score | 0.025037 (rank : 81) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.023899 (rank : 82) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.023538 (rank : 83) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.023339 (rank : 84) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.023216 (rank : 85) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
EXOC1_HUMAN
|
||||||
| NC score | 0.023011 (rank : 86) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NV70, Q8WUE7, Q96T15, Q9NZE4 | Gene names | EXOC1, SEC3, SEC3L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.022896 (rank : 87) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.022270 (rank : 88) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.021626 (rank : 89) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.021448 (rank : 90) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.021171 (rank : 91) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.021137 (rank : 92) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.021107 (rank : 93) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.021036 (rank : 94) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.020704 (rank : 95) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
KRHB5_HUMAN
|
||||||
| NC score | 0.020054 (rank : 96) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P78386, Q9NSB1 | Gene names | KRTHB5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb5 (Hair keratin, type II Hb5). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.019921 (rank : 97) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ANGL5_HUMAN
|
||||||
| NC score | 0.019654 (rank : 98) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XS5, Q86VR9 | Gene names | ANGPTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 5 precursor (Angiopoietin-like 5). | |||||
|
CN092_HUMAN
|
||||||
| NC score | 0.019418 (rank : 99) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94842 | Gene names | C14orf92, KIAA0737 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
K1C10_HUMAN
|
||||||
| NC score | 0.017881 (rank : 100) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P13645, Q14664, Q8N175 | Gene names | KRT10 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 10 (Cytokeratin-10) (CK-10) (Keratin-10) (K10). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.017478 (rank : 101) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.016545 (rank : 102) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.016530 (rank : 103) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.016023 (rank : 104) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
KRHB3_HUMAN
|
||||||
| NC score | 0.015914 (rank : 105) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
AB1IP_MOUSE
|
||||||
| NC score | 0.015543 (rank : 106) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.015272 (rank : 107) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
KRHB1_HUMAN
|
||||||
| NC score | 0.014855 (rank : 108) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
KRHB6_HUMAN
|
||||||
| NC score | 0.014851 (rank : 109) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
DAAM2_HUMAN
|
||||||
| NC score | 0.014238 (rank : 110) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
FOXP4_MOUSE
|
||||||
| NC score | 0.013855 (rank : 111) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
LMBL2_MOUSE
|
||||||
| NC score | 0.013703 (rank : 112) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59178 | Gene names | L3mbtl2 | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like 2 protein (L(3)mbt-like 2 protein) (H-l(3)mbt-like protein). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.012827 (rank : 113) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
FOXP4_HUMAN
|
||||||
| NC score | 0.011771 (rank : 114) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
RMD5B_MOUSE
|
||||||
| NC score | 0.011647 (rank : 115) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YQ7 | Gene names | Rmnd5b | |||
|
Domain Architecture |

|
|||||
| Description | RMD5 homolog B. | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.011279 (rank : 116) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
NICN1_HUMAN
|
||||||
| NC score | 0.010895 (rank : 117) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSH3, Q8IZQ2 | Gene names | NICN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicolin-1 (Tubulin polyglutamylase complex subunit 5) (PGs5) (NPCEDRG). | |||||
|
TNR25_HUMAN
|
||||||
| NC score | 0.010549 (rank : 118) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
TLK2_HUMAN
|
||||||
| NC score | 0.009796 (rank : 119) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
TLK2_MOUSE
|
||||||
| NC score | 0.009696 (rank : 120) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55047, Q9D5Y5 | Gene names | Tlk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.009499 (rank : 121) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
TLK1_HUMAN
|
||||||
| NC score | 0.008777 (rank : 122) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UKI8, Q14150, Q8N591, Q9NYH2, Q9Y4F6 | Gene names | TLK1, KIAA0137 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1) (PKU-beta). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.008301 (rank : 123) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TLK1_MOUSE
|
||||||
| NC score | 0.008027 (rank : 124) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8C0V0 | Gene names | Tlk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.007484 (rank : 125) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CAN7_HUMAN
|
||||||
| NC score | 0.005793 (rank : 126) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6W3 | Gene names | CAPN7, PALBH | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-7 (EC 3.4.22.-) (PalB homolog) (PalBH). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.005212 (rank : 127) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.005177 (rank : 128) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
SEPT2_MOUSE
|
||||||
| NC score | 0.004612 (rank : 129) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42208, Q3U9Y5 | Gene names | Sept2, Nedd-5, Nedd5 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-2 (Protein NEDD5) (Neural precursor cell expressed developmentally down-regulated protein 5). | |||||
|
SEPT2_HUMAN
|
||||||
| NC score | 0.004606 (rank : 130) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15019, Q14132, Q8IUK9, Q96CB0 | Gene names | SEPT2, DIFF6, KIAA0158, NEDD5 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-2 (Protein NEDD5). | |||||
|
FAK2_MOUSE
|
||||||
| NC score | 0.004027 (rank : 131) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
SSH1_MOUSE
|
||||||
| NC score | 0.003892 (rank : 132) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
FAK2_HUMAN
|
||||||
| NC score | 0.003750 (rank : 133) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.003361 (rank : 134) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTPRS_HUMAN
|
||||||
| NC score | 0.000595 (rank : 135) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | 0.000415 (rank : 136) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | -0.000853 (rank : 137) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||