Please be patient as the page loads
|
ARMC4_HUMAN
|
||||||
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ARMC4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
PLAK_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 2) | NC score | 0.541346 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
PLAK_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 3) | NC score | 0.525006 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
CTNB1_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 4) | NC score | 0.542489 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 5) | NC score | 0.542554 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
IMA2_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 6) | NC score | 0.323748 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |
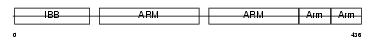
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA2_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 7) | NC score | 0.327542 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |
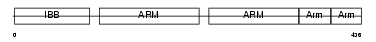
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
ARVC_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 8) | NC score | 0.405117 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
ARVC_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 9) | NC score | 0.406518 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 10) | NC score | 0.381228 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 11) | NC score | 0.331200 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
IMA5_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 12) | NC score | 0.286632 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
GDS1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 13) | NC score | 0.358632 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 14) | NC score | 0.378102 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
CTND1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 15) | NC score | 0.376962 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 16) | NC score | 0.379171 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
SPAG6_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 17) | NC score | 0.315536 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 18) | NC score | 0.370310 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
PKP4_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 19) | NC score | 0.365895 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
PKP3_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 20) | NC score | 0.337480 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 21) | NC score | 0.340242 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
RTDR1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 22) | NC score | 0.210093 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
ARMC6_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.141172 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
IMA7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.246506 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |
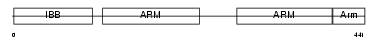
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
ARMC7_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.183150 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H6L4 | Gene names | ARMC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
PKP1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.315721 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
ARMX3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.109800 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
PKP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.319937 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
PKP2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.307080 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
RIF1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.047206 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
IMA3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.232022 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |
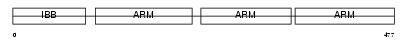
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
ARMC7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.161610 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UJZ3, Q8R599 | Gene names | Armc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
GGA1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.041421 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.041126 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
IMA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.230273 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |
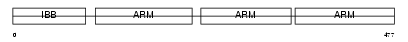
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.238076 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |
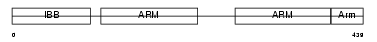
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.037523 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.021913 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
KIFA3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.062416 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70188, P70189 | Gene names | Kifap3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (KAP3). | |||||
|
K0146_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.040393 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
KIFA3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.060566 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92845, Q8NHU7, Q9H416 | Gene names | KIFAP3, SMAP | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (Smg GDS-associated protein). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.018004 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.018374 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.070904 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.067156 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.035626 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
TTC12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.036587 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
2AAB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.035873 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TNP2 | Gene names | Ppp2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
APC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.133098 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
APC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.136005 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
ARMX3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.100546 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
IMA4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.233555 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |
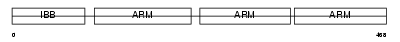
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
SART3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.018343 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLI8, Q6ZQI2, Q8BPK9, Q8C3B7, Q8CFU9 | Gene names | Sart3, Kiaa0156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (mSART-3) (Tumor-rejection antigen SART3). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.015831 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.014984 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
AP2B1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.036830 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.036832 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
ARMX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.071304 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
ARMX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.068151 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.012071 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
IMA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.226744 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
IMA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.226186 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
IMA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.232719 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |
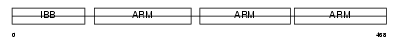
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
SARM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.053662 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
SARM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.052982 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
2AAB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.027404 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30154, O75620 | Gene names | PPP2R1B | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
AP1B1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.035545 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP1B1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.035676 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.008348 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.013922 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
LHFP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.020731 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BM86, Q3T9G7, Q3TWU2 | Gene names | Lhfp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma HMGIC fusion partner precursor. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.009867 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
RIC8B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.028187 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NVN3, Q4G103, Q6ZRN4, Q86WD3 | Gene names | RIC8B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synembryn-B (Brain synembrin) (hSyn) (Protein Ric-8B). | |||||
|
RIC8B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.027933 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80XE1, Q58L65 | Gene names | Ric8b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synembryn-B (Protein Ric-8B). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.002191 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.001999 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.027857 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.042511 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
ACOX3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.014030 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.004192 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
NCKX6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.018036 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6J4K2, Q4KMS9, Q6J4K1, Q9H6I8 | Gene names | SLC24A6, NCKX6, NCLX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium/calcium exchanger 6 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 6) (Solute carrier family 24 member 6). | |||||
|
WBP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.021419 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
ARMC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.092125 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.006397 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
COA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.010715 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
GP158_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.012730 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.016425 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.009242 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
CTNB1_MOUSE
|
||||||
| NC score | 0.542554 (rank : 2) | θ value | 7.84624e-12 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_HUMAN
|
||||||
| NC score | 0.542489 (rank : 3) | θ value | 7.84624e-12 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
PLAK_MOUSE
|
||||||
| NC score | 0.541346 (rank : 4) | θ value | 5.43371e-13 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
PLAK_HUMAN
|
||||||
| NC score | 0.525006 (rank : 5) | θ value | 1.2105e-12 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
ARVC_MOUSE
|
||||||
| NC score | 0.406518 (rank : 6) | θ value | 7.59969e-07 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.405117 (rank : 7) | θ value | 5.81887e-07 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.381228 (rank : 8) | θ value | 1.29631e-06 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.379171 (rank : 9) | θ value | 9.29e-05 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.378102 (rank : 10) | θ value | 4.1701e-05 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
CTND1_MOUSE
|
||||||
| NC score | 0.376962 (rank : 11) | θ value | 7.1131e-05 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.370310 (rank : 12) | θ value | 0.00228821 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
PKP4_HUMAN
|
||||||
| NC score | 0.365895 (rank : 13) | θ value | 0.00298849 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
GDS1_HUMAN
|
||||||
| NC score | 0.358632 (rank : 14) | θ value | 4.1701e-05 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
PKP3_HUMAN
|
||||||
| NC score | 0.340242 (rank : 15) | θ value | 0.0113563 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP3_MOUSE
|
||||||
| NC score | 0.337480 (rank : 16) | θ value | 0.00869519 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.331200 (rank : 17) | θ value | 1.09739e-05 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
IMA2_HUMAN
|
||||||
| NC score | 0.327542 (rank : 18) | θ value | 1.17247e-07 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA2_MOUSE
|
||||||
| NC score | 0.323748 (rank : 19) | θ value | 2.36244e-08 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
PKP1_MOUSE
|
||||||
| NC score | 0.319937 (rank : 20) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
PKP1_HUMAN
|
||||||
| NC score | 0.315721 (rank : 21) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
SPAG6_MOUSE
|
||||||
| NC score | 0.315536 (rank : 22) | θ value | 9.29e-05 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
PKP2_HUMAN
|
||||||
| NC score | 0.307080 (rank : 23) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
IMA5_HUMAN
|
||||||
| NC score | 0.286632 (rank : 24) | θ value | 1.43324e-05 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
IMA7_HUMAN
|
||||||
| NC score | 0.246506 (rank : 25) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
IMA7_MOUSE
|
||||||
| NC score | 0.238076 (rank : 26) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
IMA4_MOUSE
|
||||||
| NC score | 0.233555 (rank : 27) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
IMA4_HUMAN
|
||||||
| NC score | 0.232719 (rank : 28) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA3_MOUSE
|
||||||
| NC score | 0.232022 (rank : 29) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
IMA3_HUMAN
|
||||||
| NC score | 0.230273 (rank : 30) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA1_HUMAN
|
||||||
| NC score | 0.226744 (rank : 31) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
IMA1_MOUSE
|
||||||
| NC score | 0.226186 (rank : 32) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
RTDR1_HUMAN
|
||||||
| NC score | 0.210093 (rank : 33) | θ value | 0.0148317 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
ARMC7_HUMAN
|
||||||
| NC score | 0.183150 (rank : 34) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H6L4 | Gene names | ARMC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
ARMC7_MOUSE
|
||||||
| NC score | 0.161610 (rank : 35) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UJZ3, Q8R599 | Gene names | Armc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
ARMC6_MOUSE
|
||||||
| NC score | 0.141172 (rank : 36) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.136005 (rank : 37) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.133098 (rank : 38) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
ARMX3_HUMAN
|
||||||
| NC score | 0.109800 (rank : 39) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
ARMX3_MOUSE
|
||||||
| NC score | 0.100546 (rank : 40) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMC6_HUMAN
|
||||||
| NC score | 0.092125 (rank : 41) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
ARMX1_HUMAN
|
||||||
| NC score | 0.071304 (rank : 42) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.070904 (rank : 43) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX1_MOUSE
|
||||||
| NC score | 0.068151 (rank : 44) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.067156 (rank : 45) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
KIFA3_MOUSE
|
||||||
| NC score | 0.062416 (rank : 46) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70188, P70189 | Gene names | Kifap3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (KAP3). | |||||
|
KIFA3_HUMAN
|
||||||
| NC score | 0.060566 (rank : 47) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92845, Q8NHU7, Q9H416 | Gene names | KIFAP3, SMAP | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-associated protein 3 (Smg GDS-associated protein). | |||||
|
SARM1_HUMAN
|
||||||
| NC score | 0.053662 (rank : 48) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
SARM1_MOUSE
|
||||||
| NC score | 0.052982 (rank : 49) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
RIF1_HUMAN
|
||||||
| NC score | 0.047206 (rank : 50) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.042511 (rank : 51) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.041421 (rank : 52) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.041126 (rank : 53) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
K0146_MOUSE
|
||||||
| NC score | 0.040393 (rank : 54) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.037523 (rank : 55) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
AP2B1_MOUSE
|
||||||
| NC score | 0.036832 (rank : 56) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_HUMAN
|
||||||
| NC score | 0.036830 (rank : 57) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.036587 (rank : 58) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
2AAB_MOUSE
|
||||||
| NC score | 0.035873 (rank : 59) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TNP2 | Gene names | Ppp2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
AP1B1_MOUSE
|
||||||
| NC score | 0.035676 (rank : 60) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.035626 (rank : 61) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
AP1B1_HUMAN
|
||||||
| NC score | 0.035545 (rank : 62) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
RIC8B_HUMAN
|
||||||
| NC score | 0.028187 (rank : 63) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NVN3, Q4G103, Q6ZRN4, Q86WD3 | Gene names | RIC8B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synembryn-B (Brain synembrin) (hSyn) (Protein Ric-8B). | |||||
|
RIC8B_MOUSE
|
||||||
| NC score | 0.027933 (rank : 64) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80XE1, Q58L65 | Gene names | Ric8b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synembryn-B (Protein Ric-8B). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.027857 (rank : 65) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
2AAB_HUMAN
|
||||||
| NC score | 0.027404 (rank : 66) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30154, O75620 | Gene names | PPP2R1B | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.021913 (rank : 67) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
WBP4_MOUSE
|
||||||
| NC score | 0.021419 (rank : 68) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
LHFP_MOUSE
|
||||||
| NC score | 0.020731 (rank : 69) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BM86, Q3T9G7, Q3TWU2 | Gene names | Lhfp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma HMGIC fusion partner precursor. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.018374 (rank : 70) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SART3_MOUSE
|
||||||
| NC score | 0.018343 (rank : 71) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLI8, Q6ZQI2, Q8BPK9, Q8C3B7, Q8CFU9 | Gene names | Sart3, Kiaa0156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Squamous cell carcinoma antigen recognized by T-cells 3 (SART-3) (mSART-3) (Tumor-rejection antigen SART3). | |||||
|
NCKX6_HUMAN
|
||||||
| NC score | 0.018036 (rank : 72) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6J4K2, Q4KMS9, Q6J4K1, Q9H6I8 | Gene names | SLC24A6, NCKX6, NCLX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium/calcium exchanger 6 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 6) (Solute carrier family 24 member 6). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.018004 (rank : 73) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.016425 (rank : 74) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.015831 (rank : 75) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.014984 (rank : 76) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
ACOX3_MOUSE
|
||||||
| NC score | 0.014030 (rank : 77) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPL9 | Gene names | Acox3 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 3, peroxisomal (EC 1.3.3.6) (Pristanoyl-CoA oxidase) (Branched-chain acyl-CoA oxidase) (BRCACox). | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.013922 (rank : 78) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.012730 (rank : 79) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.012071 (rank : 80) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.010715 (rank : 81) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.009867 (rank : 82) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.009242 (rank : 83) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.008348 (rank : 84) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.006397 (rank : 85) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.004192 (rank : 86) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.002191 (rank : 87) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.001999 (rank : 88) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||