Please be patient as the page loads
|
AP2B1_HUMAN
|
||||||
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AP1B1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997125 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP1B1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996343 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP2B1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999941 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP4B1_MOUSE
|
||||||
| θ value | 5.15425e-80 (rank : 5) | NC score | 0.891964 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WV76 | Gene names | Ap4b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta-1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP4B1_HUMAN
|
||||||
| θ value | 3.69378e-78 (rank : 6) | NC score | 0.885518 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y6B7 | Gene names | AP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta 1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 4.67263e-73 (rank : 7) | NC score | 0.806442 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 5.1662e-72 (rank : 8) | NC score | 0.805576 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 6.7473e-72 (rank : 9) | NC score | 0.801767 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 4.37348e-71 (rank : 10) | NC score | 0.803242 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 11) | NC score | 0.500624 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 12) | NC score | 0.499275 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 13) | NC score | 0.575650 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP2A1_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 14) | NC score | 0.473435 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 15) | NC score | 0.476148 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 16) | NC score | 0.551432 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 17) | NC score | 0.509262 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 18) | NC score | 0.510242 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
COPB_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 19) | NC score | 0.532967 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 20) | NC score | 0.532808 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPG2_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 21) | NC score | 0.412281 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG2_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 22) | NC score | 0.408062 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 23) | NC score | 0.370401 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 24) | NC score | 0.500511 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP4E1_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 25) | NC score | 0.458027 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP4E1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 26) | NC score | 0.453651 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 27) | NC score | 0.455225 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
SPAG6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 28) | NC score | 0.086042 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
K0056_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.086051 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.097830 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
EVPL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.022008 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.030124 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.032582 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
K0056_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.080319 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
TRIM7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.025767 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
RRP5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.051549 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
PON2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.022793 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15165, O15114, O15115, O75856, Q86YL0 | Gene names | PON2 | |||
|
Domain Architecture |

|
|||||
| Description | Serum paraoxonase/arylesterase 2 (EC 3.1.1.2) (EC 3.1.8.1) (PON 2) (Serum aryldialkylphosphatase 2) (A-esterase 2) (Aromatic esterase 2). | |||||
|
RPTOR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.049339 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N122, Q8N4V9, Q8TB32, Q9P2P3 | Gene names | RAPTOR, KIAA1303 | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.015007 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.014786 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TRIM7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.017911 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923T7, Q5NCB6, Q99PQ5 | Gene names | Trim7, Gnip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein). | |||||
|
UB2R1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.017928 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49427 | Gene names | CDC34, UBE2R1 | |||
|
Domain Architecture |
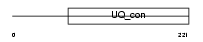
|
|||||
| Description | Ubiquitin-conjugating enzyme E2-32 kDa complementing (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein) (E2-CDC34). | |||||
|
UB2R1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.017921 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFI2, Q505K8 | Gene names | Cdc34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-32 kDa complementing (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein) (E2-CDC34). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.019156 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
RPTOR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.044601 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4Q0, Q8C9W9, Q8CBY4, Q8CDY8, Q9D4H3 | Gene names | Raptor | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.036830 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | -0.000397 (rank : 61) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
DOCK8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.009206 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.049631 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.003451 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.006153 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
IL5RA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.012670 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01344, Q6ISX9 | Gene names | IL5RA, IL5R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-5 receptor alpha chain precursor (IL-5R-alpha) (CD125 antigen) (CDw125). | |||||
|
NF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.015840 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21359, O00662, Q14284, Q14930, Q9UMK3 | Gene names | NF1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1) [Contains: Neurofibromin truncated]. | |||||
|
NF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.015791 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04690, Q61956, Q61957 | Gene names | Nf1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1). | |||||
|
PK3CA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.014139 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.002602 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
IPO4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.009213 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.001102 (rank : 60) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
PK3CA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.012527 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.051357 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050943 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AP2B1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_MOUSE
|
||||||
| NC score | 0.999941 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP1B1_HUMAN
|
||||||
| NC score | 0.997125 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP1B1_MOUSE
|
||||||
| NC score | 0.996343 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AP4B1_MOUSE
|
||||||
| NC score | 0.891964 (rank : 5) | θ value | 5.15425e-80 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WV76 | Gene names | Ap4b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta-1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP4B1_HUMAN
|
||||||
| NC score | 0.885518 (rank : 6) | θ value | 3.69378e-78 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y6B7 | Gene names | AP4B1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit beta-1 (Adapter-related protein complex 4 beta 1 subunit) (Beta subunit of AP-4) (AP-4 adapter complex beta subunit). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.806442 (rank : 7) | θ value | 4.67263e-73 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.805576 (rank : 8) | θ value | 5.1662e-72 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.803242 (rank : 9) | θ value | 4.37348e-71 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.801767 (rank : 10) | θ value | 6.7473e-72 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.575650 (rank : 11) | θ value | 2.13179e-17 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.551432 (rank : 12) | θ value | 1.16975e-15 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
COPB_HUMAN
|
||||||
| NC score | 0.532967 (rank : 13) | θ value | 4.59992e-12 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53618, Q6GTT7, Q9NTK2, Q9UNW7 | Gene names | COPB1, COPB | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
COPB_MOUSE
|
||||||
| NC score | 0.532808 (rank : 14) | θ value | 4.59992e-12 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JIF7 | Gene names | Copb1, Copb | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit beta (Beta-coat protein) (Beta-COP). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.510242 (rank : 15) | θ value | 4.16044e-13 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.509262 (rank : 16) | θ value | 4.16044e-13 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.500624 (rank : 17) | θ value | 1.02238e-19 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.500511 (rank : 18) | θ value | 2.13673e-09 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP2A2_MOUSE
|
||||||
| NC score | 0.499275 (rank : 19) | θ value | 2.27762e-19 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.476148 (rank : 20) | θ value | 8.10077e-17 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP2A1_MOUSE
|
||||||
| NC score | 0.473435 (rank : 21) | θ value | 6.20254e-17 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
AP4E1_HUMAN
|
||||||
| NC score | 0.458027 (rank : 22) | θ value | 8.97725e-08 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPM8, Q9Y588 | Gene names | AP4E1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.455225 (rank : 23) | θ value | 3.41135e-07 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP4E1_MOUSE
|
||||||
| NC score | 0.453651 (rank : 24) | θ value | 2.61198e-07 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80V94 | Gene names | Ap4e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-4 complex subunit epsilon-1 (Adapter-related protein complex 4 epsilon-1 subunit) (Epsilon subunit of AP-4) (AP-4 adapter complex epsilon subunit). | |||||
|
COPG2_MOUSE
|
||||||
| NC score | 0.412281 (rank : 25) | θ value | 1.133e-10 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG2_HUMAN
|
||||||
| NC score | 0.408062 (rank : 26) | θ value | 1.47974e-10 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG_HUMAN
|
||||||
| NC score | 0.370401 (rank : 27) | θ value | 1.25267e-09 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.097830 (rank : 28) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
K0056_MOUSE
|
||||||
| NC score | 0.086051 (rank : 29) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
SPAG6_MOUSE
|
||||||
| NC score | 0.086042 (rank : 30) | θ value | 0.0961366 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
K0056_HUMAN
|
||||||
| NC score | 0.080319 (rank : 31) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.051549 (rank : 32) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.051357 (rank : 33) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.050943 (rank : 34) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.049631 (rank : 35) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
RPTOR_HUMAN
|
||||||
| NC score | 0.049339 (rank : 36) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N122, Q8N4V9, Q8TB32, Q9P2P3 | Gene names | RAPTOR, KIAA1303 | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
RPTOR_MOUSE
|
||||||
| NC score | 0.044601 (rank : 37) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4Q0, Q8C9W9, Q8CBY4, Q8CDY8, Q9D4H3 | Gene names | Raptor | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.036830 (rank : 38) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.032582 (rank : 39) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.030124 (rank : 40) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
TRIM7_HUMAN
|
||||||
| NC score | 0.025767 (rank : 41) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
PON2_HUMAN
|
||||||
| NC score | 0.022793 (rank : 42) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15165, O15114, O15115, O75856, Q86YL0 | Gene names | PON2 | |||
|
Domain Architecture |

|
|||||
| Description | Serum paraoxonase/arylesterase 2 (EC 3.1.1.2) (EC 3.1.8.1) (PON 2) (Serum aryldialkylphosphatase 2) (A-esterase 2) (Aromatic esterase 2). | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.022008 (rank : 43) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.019156 (rank : 44) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
UB2R1_HUMAN
|
||||||
| NC score | 0.017928 (rank : 45) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49427 | Gene names | CDC34, UBE2R1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-32 kDa complementing (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein) (E2-CDC34). | |||||
|
UB2R1_MOUSE
|
||||||
| NC score | 0.017921 (rank : 46) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFI2, Q505K8 | Gene names | Cdc34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2-32 kDa complementing (EC 6.3.2.19) (Ubiquitin-protein ligase) (Ubiquitin carrier protein) (E2-CDC34). | |||||
|
TRIM7_MOUSE
|
||||||
| NC score | 0.017911 (rank : 47) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923T7, Q5NCB6, Q99PQ5 | Gene names | Trim7, Gnip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein). | |||||
|
NF1_HUMAN
|
||||||
| NC score | 0.015840 (rank : 48) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21359, O00662, Q14284, Q14930, Q9UMK3 | Gene names | NF1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1) [Contains: Neurofibromin truncated]. | |||||
|
NF1_MOUSE
|
||||||
| NC score | 0.015791 (rank : 49) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04690, Q61956, Q61957 | Gene names | Nf1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.015007 (rank : 50) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.014786 (rank : 51) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
PK3CA_MOUSE
|
||||||
| NC score | 0.014139 (rank : 52) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
IL5RA_HUMAN
|
||||||
| NC score | 0.012670 (rank : 53) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01344, Q6ISX9 | Gene names | IL5RA, IL5R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-5 receptor alpha chain precursor (IL-5R-alpha) (CD125 antigen) (CDw125). | |||||
|
PK3CA_HUMAN
|
||||||
| NC score | 0.012527 (rank : 54) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
IPO4_MOUSE
|
||||||
| NC score | 0.009213 (rank : 55) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
DOCK8_HUMAN
|
||||||
| NC score | 0.009206 (rank : 56) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NF50, Q3MV16, Q5JPJ1, Q8TEP1, Q8WUY2, Q9BYJ5, Q9H1Q2, Q9H1Q3, Q9H308, Q9H7P2 | Gene names | DOCK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dedicator of cytokinesis protein 8. | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.006153 (rank : 57) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.003451 (rank : 58) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.002602 (rank : 59) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.001102 (rank : 60) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | -0.000397 (rank : 61) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||