Please be patient as the page loads
|
NF1_HUMAN
|
||||||
| SwissProt Accessions | P21359, O00662, Q14284, Q14930, Q9UMK3 | Gene names | NF1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1) [Contains: Neurofibromin truncated]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P21359, O00662, Q14284, Q14930, Q9UMK3 | Gene names | NF1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1) [Contains: Neurofibromin truncated]. | |||||
|
NF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998950 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q04690, Q61956, Q61957 | Gene names | Nf1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1). | |||||
|
DAB2P_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 3) | NC score | 0.592489 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
DAB2P_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 4) | NC score | 0.591870 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
RASA1_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 5) | NC score | 0.477324 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
NGAP_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 6) | NC score | 0.556901 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
RASA3_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 7) | NC score | 0.508070 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 8) | NC score | 0.521213 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
RASA3_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 9) | NC score | 0.510647 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
RASA2_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 10) | NC score | 0.495659 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA2_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 11) | NC score | 0.483365 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASL1_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 12) | NC score | 0.443546 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL1_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 13) | NC score | 0.455162 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL2_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 14) | NC score | 0.407250 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
IQGA1_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 15) | NC score | 0.357322 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
IQGA2_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 16) | NC score | 0.383320 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
IQGA1_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 17) | NC score | 0.346858 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
IQGA3_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 18) | NC score | 0.342416 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||
|
RHG08_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.052934 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
ABCAD_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.021618 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
PDE4C_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.025729 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.037935 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.023997 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.022825 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.019903 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.031715 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
ECE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.025425 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42892, Q14217, Q58GE7, Q5THM5, Q5THM7, Q5THM8, Q9UJQ6, Q9UPF4, Q9UPM4, Q9Y501 | Gene names | ECE1 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme 1 (EC 3.4.24.71) (ECE-1). | |||||
|
K0133_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.049105 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14146 | Gene names | KIAA0133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0133. | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.013123 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.019470 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
ARMC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.039076 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
PDE4C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.019054 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
ZN343_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | -0.000407 (rank : 65) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1L6, Q5JXU8, Q5JXU9, Q8N8F2, Q96EX8, Q96NA5, Q9BQB0 | Gene names | ZNF343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 343. | |||||
|
ARPC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.046150 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15144, Q92801, Q9P1D4 | Gene names | ARPC2, ARC34 | |||
|
Domain Architecture |
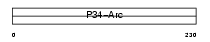
|
|||||
| Description | Actin-related protein 2/3 complex subunit 2 (ARP2/3 complex 34 kDa subunit) (p34-ARC). | |||||
|
MMEL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.021961 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLI3, Q3U495, Q9ERK2, Q9ERK3, Q9QZV6, Q9QZV7 | Gene names | Mmel1, Nep2, Nl1, Sep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane metallo-endopeptidase-like 1 (EC 3.4.24.11) (Neprilysin-2) (Neprilysin II) (NL2) (NEPII) (NEP2(m)) (Neprilysin-like peptidase) (NEPLP) (Neprilysin-like 1) (NL-1) (Soluble secreted endopeptidase) [Contains: Membrane metallo-endopeptidase-like 1, soluble form (Neprilysin-2 secreted) (NEP2(s))]. | |||||
|
NELFB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.034135 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4Y3, Q99J41, Q9JJA5 | Gene names | Nelfb | |||
|
Domain Architecture |

|
|||||
| Description | Negative elongation factor B (NELF-B). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.014183 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
TLR5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.012513 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLF7 | Gene names | Tlr5 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 5 precursor. | |||||
|
ARPC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.042663 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CVB6, Q3U870 | Gene names | Arpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-related protein 2/3 complex subunit 2 (ARP2/3 complex 34 kDa subunit) (p34-ARC). | |||||
|
DMXL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.017478 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
AMPE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.014488 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07075 | Gene names | ENPEP | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). | |||||
|
CP39A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.012512 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |
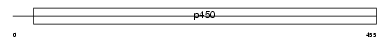
|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
TIM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.024347 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1X4, Q63ZX9, Q6P204, Q6PDL4, Q7TPV8, Q8R0Q2, Q9R268, Q9Z0E7 | Gene names | Timeless, Tim1, Timeless1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Timeless homolog (mTim). | |||||
|
AMPE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.013254 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16406 | Gene names | Enpep | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (BP-1/6C3 antigen). | |||||
|
AP2B1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.015840 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.015842 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.014560 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
AP1B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.014138 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
CAND1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.014742 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86VP6, O94918, Q6PIY4, Q8NDJ4, Q96JZ9, Q96T19, Q9BTC4, Q9H0G2, Q9P0H7, Q9UF85 | Gene names | CAND1, KIAA0829, TIP120, TIP120A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1) (TBP-interacting protein TIP120A) (TBP-interacting protein of 120 kDa A). | |||||
|
CAND1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.014769 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ38, Q6PFR0, Q9CV45 | Gene names | Cand1, D10Ertd516e, Kiaa0829 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1). | |||||
|
ECE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.017060 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q4PZA2, Q4PZ99, Q4PZA1, Q6P9Q9 | Gene names | Ece1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelin-converting enzyme 1 (EC 3.4.24.71) (ECE-1). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.012261 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.008717 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
K1HA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.001308 (rank : 64) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76009 | Gene names | KRTHA3A, HHA3-I, HKA3A | |||
|
Domain Architecture |
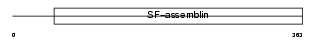
|
|||||
| Description | Keratin, type I cuticular Ha3-I (Hair keratin, type I Ha3-I). | |||||
|
KCTD3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.007389 (rank : 63) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
TOP2B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.010542 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64511, Q7TQG4 | Gene names | Top2b | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
FA62A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.059557 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.067222 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
PKHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.059174 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.061208 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
PKHA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.052021 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
RFIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.063619 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
SYT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.050234 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.050465 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.051936 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |
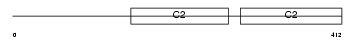
|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
NF1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P21359, O00662, Q14284, Q14930, Q9UMK3 | Gene names | NF1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1) [Contains: Neurofibromin truncated]. | |||||
|
NF1_MOUSE
|
||||||
| NC score | 0.998950 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q04690, Q61956, Q61957 | Gene names | Nf1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1). | |||||
|
DAB2P_MOUSE
|
||||||
| NC score | 0.592489 (rank : 3) | θ value | 3.39556e-23 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
DAB2P_HUMAN
|
||||||
| NC score | 0.591870 (rank : 4) | θ value | 4.43474e-23 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
NGAP_HUMAN
|
||||||
| NC score | 0.556901 (rank : 5) | θ value | 3.75424e-22 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.521213 (rank : 6) | θ value | 2.5182e-18 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
RASA3_MOUSE
|
||||||
| NC score | 0.510647 (rank : 7) | θ value | 2.13179e-17 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
RASA3_HUMAN
|
||||||
| NC score | 0.508070 (rank : 8) | θ value | 2.06002e-20 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
RASA2_HUMAN
|
||||||
| NC score | 0.495659 (rank : 9) | θ value | 2.7842e-17 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.483365 (rank : 10) | θ value | 2.35696e-16 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA1_HUMAN
|
||||||
| NC score | 0.477324 (rank : 11) | θ value | 1.6852e-22 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
RASL1_MOUSE
|
||||||
| NC score | 0.455162 (rank : 12) | θ value | 1.2105e-12 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL1_HUMAN
|
||||||
| NC score | 0.443546 (rank : 13) | θ value | 5.43371e-13 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL2_HUMAN
|
||||||
| NC score | 0.407250 (rank : 14) | θ value | 3.52202e-12 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
IQGA2_HUMAN
|
||||||
| NC score | 0.383320 (rank : 15) | θ value | 5.08577e-11 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
IQGA1_MOUSE
|
||||||
| NC score | 0.357322 (rank : 16) | θ value | 2.98157e-11 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
IQGA1_HUMAN
|
||||||
| NC score | 0.346858 (rank : 17) | θ value | 1.133e-10 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P46940 | Gene names | IQGAP1, KIAA0051 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1 (p195). | |||||
|
IQGA3_HUMAN
|
||||||
| NC score | 0.342416 (rank : 18) | θ value | 1.06045e-08 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.067222 (rank : 19) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
RFIP2_HUMAN
|
||||||
| NC score | 0.063619 (rank : 20) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
PKHA1_MOUSE
|
||||||
| NC score | 0.061208 (rank : 21) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BUL6, Q8BK96, Q8BXU1, Q8VE13 | Gene names | Plekha1, Tapp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.059557 (rank : 22) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
PKHA1_HUMAN
|
||||||
| NC score | 0.059174 (rank : 23) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HB21, Q9BVK0 | Gene names | PLEKHA1, TAPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 1 (Tandem PH domain-containing protein 1) (TAPP-1). | |||||
|
RHG08_MOUSE
|
||||||
| NC score | 0.052934 (rank : 24) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CXP4, Q99JY7 | Gene names | Arhgap8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
PKHA2_MOUSE
|
||||||
| NC score | 0.052021 (rank : 25) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ERS5, Q8BY29 | Gene names | Plekha2, Tapp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 2 (Tandem PH domain-containing protein 2) (TAPP-2) (PH domain-containing adaptor PHAD47). | |||||
|
SYT7_HUMAN
|
||||||
| NC score | 0.051936 (rank : 26) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT3_MOUSE
|
||||||
| NC score | 0.050465 (rank : 27) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT3_HUMAN
|
||||||
| NC score | 0.050234 (rank : 28) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
K0133_HUMAN
|
||||||
| NC score | 0.049105 (rank : 29) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14146 | Gene names | KIAA0133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0133. | |||||
|
ARPC2_HUMAN
|
||||||
| NC score | 0.046150 (rank : 30) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15144, Q92801, Q9P1D4 | Gene names | ARPC2, ARC34 | |||
|
Domain Architecture |

|
|||||
| Description | Actin-related protein 2/3 complex subunit 2 (ARP2/3 complex 34 kDa subunit) (p34-ARC). | |||||
|
ARPC2_MOUSE
|
||||||
| NC score | 0.042663 (rank : 31) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CVB6, Q3U870 | Gene names | Arpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-related protein 2/3 complex subunit 2 (ARP2/3 complex 34 kDa subunit) (p34-ARC). | |||||
|
ARMC6_HUMAN
|
||||||
| NC score | 0.039076 (rank : 32) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.037935 (rank : 33) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NELFB_MOUSE
|
||||||
| NC score | 0.034135 (rank : 34) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4Y3, Q99J41, Q9JJA5 | Gene names | Nelfb | |||
|
Domain Architecture |

|
|||||
| Description | Negative elongation factor B (NELF-B). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.031715 (rank : 35) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
PDE4C_MOUSE
|
||||||
| NC score | 0.025729 (rank : 36) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UEI1, Q8K0P4 | Gene names | Pde4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17). | |||||
|
ECE1_HUMAN
|
||||||
| NC score | 0.025425 (rank : 37) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42892, Q14217, Q58GE7, Q5THM5, Q5THM7, Q5THM8, Q9UJQ6, Q9UPF4, Q9UPM4, Q9Y501 | Gene names | ECE1 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme 1 (EC 3.4.24.71) (ECE-1). | |||||
|
TIM_MOUSE
|
||||||
| NC score | 0.024347 (rank : 38) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1X4, Q63ZX9, Q6P204, Q6PDL4, Q7TPV8, Q8R0Q2, Q9R268, Q9Z0E7 | Gene names | Timeless, Tim1, Timeless1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Timeless homolog (mTim). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.023997 (rank : 39) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.022825 (rank : 40) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
MMEL1_MOUSE
|
||||||
| NC score | 0.021961 (rank : 41) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLI3, Q3U495, Q9ERK2, Q9ERK3, Q9QZV6, Q9QZV7 | Gene names | Mmel1, Nep2, Nl1, Sep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane metallo-endopeptidase-like 1 (EC 3.4.24.11) (Neprilysin-2) (Neprilysin II) (NL2) (NEPII) (NEP2(m)) (Neprilysin-like peptidase) (NEPLP) (Neprilysin-like 1) (NL-1) (Soluble secreted endopeptidase) [Contains: Membrane metallo-endopeptidase-like 1, soluble form (Neprilysin-2 secreted) (NEP2(s))]. | |||||
|
ABCAD_HUMAN
|
||||||
| NC score | 0.021618 (rank : 42) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.019903 (rank : 43) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.019470 (rank : 44) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PDE4C_HUMAN
|
||||||
| NC score | 0.019054 (rank : 45) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08493, Q9UN44, Q9UN45, Q9UN46, Q9UPJ6 | Gene names | PDE4C | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4C (EC 3.1.4.17) (DPDE1) (PDE21). | |||||
|
DMXL1_MOUSE
|
||||||
| NC score | 0.017478 (rank : 46) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
ECE1_MOUSE
|
||||||
| NC score | 0.017060 (rank : 47) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q4PZA2, Q4PZ99, Q4PZA1, Q6P9Q9 | Gene names | Ece1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelin-converting enzyme 1 (EC 3.4.24.71) (ECE-1). | |||||
|
AP2B1_MOUSE
|
||||||
| NC score | 0.015842 (rank : 48) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_HUMAN
|
||||||
| NC score | 0.015840 (rank : 49) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
CAND1_MOUSE
|
||||||
| NC score | 0.014769 (rank : 50) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ38, Q6PFR0, Q9CV45 | Gene names | Cand1, D10Ertd516e, Kiaa0829 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1). | |||||
|
CAND1_HUMAN
|
||||||
| NC score | 0.014742 (rank : 51) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86VP6, O94918, Q6PIY4, Q8NDJ4, Q96JZ9, Q96T19, Q9BTC4, Q9H0G2, Q9P0H7, Q9UF85 | Gene names | CAND1, KIAA0829, TIP120, TIP120A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1) (TBP-interacting protein TIP120A) (TBP-interacting protein of 120 kDa A). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.014560 (rank : 52) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
AMPE_HUMAN
|
||||||
| NC score | 0.014488 (rank : 53) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07075 | Gene names | ENPEP | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.014183 (rank : 54) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
AP1B1_MOUSE
|
||||||
| NC score | 0.014138 (rank : 55) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
AMPE_MOUSE
|
||||||
| NC score | 0.013254 (rank : 56) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16406 | Gene names | Enpep | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (BP-1/6C3 antigen). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.013123 (rank : 57) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
TLR5_MOUSE
|
||||||
| NC score | 0.012513 (rank : 58) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLF7 | Gene names | Tlr5 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 5 precursor. | |||||
|
CP39A_HUMAN
|
||||||
| NC score | 0.012512 (rank : 59) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYL5, Q96FW5 | Gene names | CYP39A1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 39A1 (EC 1.14.13.99) (24-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (hCYP39A1). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.012261 (rank : 60) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
TOP2B_MOUSE
|
||||||
| NC score | 0.010542 (rank : 61) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64511, Q7TQG4 | Gene names | Top2b | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.008717 (rank : 62) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
KCTD3_MOUSE
|
||||||
| NC score | 0.007389 (rank : 63) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
K1HA_HUMAN
|
||||||
| NC score | 0.001308 (rank : 64) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76009 | Gene names | KRTHA3A, HHA3-I, HKA3A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha3-I (Hair keratin, type I Ha3-I). | |||||
|
ZN343_HUMAN
|
||||||
| NC score | -0.000407 (rank : 65) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1L6, Q5JXU8, Q5JXU9, Q8N8F2, Q96EX8, Q96NA5, Q9BQB0 | Gene names | ZNF343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 343. | |||||