Please be patient as the page loads
|
AMPE_HUMAN
|
||||||
| SwissProt Accessions | Q07075 | Gene names | ENPEP | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AMPE_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q07075 | Gene names | ENPEP | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). | |||||
|
AMPE_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998150 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16406 | Gene names | Enpep | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (BP-1/6C3 antigen). | |||||
|
AMPN_HUMAN
|
||||||
| θ value | 4.55495e-161 (rank : 3) | NC score | 0.977099 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15144, Q16728, Q8IUK3, Q8IVH3, Q9UCE0 | Gene names | ANPEP, APN, CD13, PEPN | |||
|
Domain Architecture |
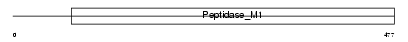
|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (hAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (gp150) (Myeloid plasma membrane glycoprotein CD13) (CD13 antigen). | |||||
|
ARTS1_MOUSE
|
||||||
| θ value | 3.49569e-153 (rank : 4) | NC score | 0.974986 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EQH2, Q9ET63 | Gene names | Arts1, Appils | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (VEGF-induced aminopeptidase). | |||||
|
AMPN_MOUSE
|
||||||
| θ value | 1.91815e-151 (rank : 5) | NC score | 0.975583 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97449, Q91YH8, Q99K63 | Gene names | Anpep, Lap-1, Lap1 | |||
|
Domain Architecture |
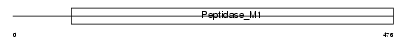
|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (mAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (Membrane protein p161) (CD13 antigen). | |||||
|
ARTS1_HUMAN
|
||||||
| θ value | 6.38546e-147 (rank : 6) | NC score | 0.974753 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZ08, O60278, Q6UWY6, Q8NEL4, Q8TAD0, Q9UHF8, Q9UKY2 | Gene names | ARTS1, APPILS, KIAA0525 | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator). | |||||
|
TRHDE_MOUSE
|
||||||
| θ value | 2.12569e-142 (rank : 7) | NC score | 0.968984 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K093 | Gene names | Trhde | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
TRHDE_HUMAN
|
||||||
| θ value | 6.18479e-142 (rank : 8) | NC score | 0.970483 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKU6, Q6UWJ4 | Gene names | TRHDE | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
PSA_MOUSE
|
||||||
| θ value | 7.8238e-137 (rank : 9) | NC score | 0.970038 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
PSA_HUMAN
|
||||||
| θ value | 2.27638e-136 (rank : 10) | NC score | 0.969990 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
LCAP_HUMAN
|
||||||
| θ value | 1.80368e-133 (rank : 11) | NC score | 0.974423 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIQ6, O00769, Q15145, Q9UIQ7 | Gene names | LNPEP, OTASE | |||
|
Domain Architecture |
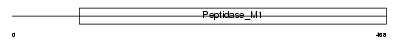
|
|||||
| Description | Leucyl-cystinyl aminopeptidase (EC 3.4.11.3) (Cystinyl aminopeptidase) (Oxytocinase) (OTase) (Insulin-regulated membrane aminopeptidase) (Insulin-responsive aminopeptidase) (IRAP) (Placental leucine aminopeptidase) (P-LAP). | |||||
|
LAEVR_HUMAN
|
||||||
| θ value | 2.14053e-118 (rank : 12) | NC score | 0.963031 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6Q4G3, Q32MR1, Q4G0I9, Q4G0V2, Q86XA3, Q8NBZ2 | Gene names | LVRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-) (CHL2 antigen). | |||||
|
LAEVR_MOUSE
|
||||||
| θ value | 7.70199e-76 (rank : 13) | NC score | 0.955472 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q2KHK3, Q9D633 | Gene names | Lvrn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-). | |||||
|
LKHA4_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 14) | NC score | 0.696002 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09960 | Gene names | LTA4H, LTA4 | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
LKHA4_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 15) | NC score | 0.688492 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24527, Q8VDR8 | Gene names | Lta4h | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
AMPB_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 16) | NC score | 0.651714 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCT3 | Gene names | Rnpep | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase) (Cytosol aminopeptidase IV). | |||||
|
AMPB_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 17) | NC score | 0.636908 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H4A4, Q9BVM9, Q9H1D4, Q9NPT7 | Gene names | RNPEP, APB | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase). | |||||
|
RNPL1_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 18) | NC score | 0.605235 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
AMPO_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 19) | NC score | 0.444341 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N6M6, Q5T9B1, Q5T9B3, Q5T9B4, Q8WUL6, Q96M23, Q96SS1 | Gene names | ONPEP, C9orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
TAF2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 20) | NC score | 0.227822 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1X5, O43487, O43604, O60668, Q86WW7, Q8IWK4 | Gene names | TAF2, CIF150, TAF2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa) (TAFII-150) (TAFII150) (150 kDa cofactor of initiator function). | |||||
|
TAF2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 21) | NC score | 0.218881 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C176, Q8BKQ7 | Gene names | Taf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa). | |||||
|
AMPO_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 22) | NC score | 0.426821 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BXQ6, Q6P0W6, Q6P394, Q8BHX5 | Gene names | Onpep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
ARHG7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.020682 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
ARHG6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.023933 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
ARHG6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.021507 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4I3, Q8C9V4 | Gene names | Arhgef6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6). | |||||
|
PACN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.020698 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UNF0, O95921, Q96HV9, Q9H0D3, Q9NPN1, Q9Y4V2 | Gene names | PACSIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
PACN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.020422 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVE8 | Gene names | Pacsin2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
CSKP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | -0.000421 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||
|
ILEU_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.003796 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30740, Q53FB9, Q5W0E1 | Gene names | SERPINB1, ELANH2, PI2 | |||
|
Domain Architecture |
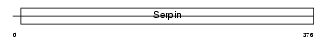
|
|||||
| Description | Leukocyte elastase inhibitor (LEI) (Serpin B1) (Monocyte/neutrophil elastase inhibitor) (M/NEI) (EI). | |||||
|
OCTC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.010583 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKG9, Q8IUW9, Q9Y6I2 | Gene names | CROT, COT | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal carnitine O-octanoyltransferase (EC 2.3.1.137) (COT). | |||||
|
RAD9A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.011082 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0F6, Q8QZZ3 | Gene names | Rad9a, Rad9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint control protein RAD9A (EC 3.1.11.2) (DNA repair exonuclease rad9 homolog A) (Rad9-like protein) (mRAD9). | |||||
|
RO60_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.012524 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08848, Q9QYD8 | Gene names | Trove2, Ssa2 | |||
|
Domain Architecture |

|
|||||
| Description | 60 kDa SS-A/Ro ribonucleoprotein (60 kDa Ro protein) (60 kDa ribonucleoprotein Ro) (RoRNP) (TROVE domain family member 2). | |||||
|
NF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.014488 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21359, O00662, Q14284, Q14930, Q9UMK3 | Gene names | NF1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1) [Contains: Neurofibromin truncated]. | |||||
|
NF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.014443 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04690, Q61956, Q61957 | Gene names | Nf1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1). | |||||
|
ARFG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.005585 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | -0.000304 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
HEP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.002746 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49182 | Gene names | Serpind1, Hcf2, Hcii | |||
|
Domain Architecture |

|
|||||
| Description | Heparin cofactor 2 precursor (Heparin cofactor II) (HC-II) (Protease inhibitor leuserpin 2). | |||||
|
KLOT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.005048 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35082, O70175, O70621 | Gene names | Kl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Klotho precursor (EC 3.2.1.31) [Contains: Klotho peptide]. | |||||
|
OBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.009780 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64693 | Gene names | Pou2af1, Obf-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain class 2-associating factor 1 (B-cell-specific coactivator OBF-1) (OCT-binding factor 1) (BOB-1) (BOB1) (OCA-B). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.002109 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
STA13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.003759 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
ECEL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.009876 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95672, Q45UD9, Q53RF9, Q6UW86, Q86TH4, Q9NY95 | Gene names | ECEL1, XCE | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein). | |||||
|
ECEL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.009874 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JMI0 | Gene names | Ecel1, Dine, Xce | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein) (Damage-induced neuronal endopeptidase). | |||||
|
OCTC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.007915 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DC50, Q921I4 | Gene names | Crot, Cot | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal carnitine O-octanoyltransferase (EC 2.3.1.137) (COT). | |||||
|
SYT12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.002162 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
TSNA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.005813 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
AMPE_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q07075 | Gene names | ENPEP | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). | |||||
|
AMPE_MOUSE
|
||||||
| NC score | 0.998150 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16406 | Gene names | Enpep | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (BP-1/6C3 antigen). | |||||
|
AMPN_HUMAN
|
||||||
| NC score | 0.977099 (rank : 3) | θ value | 4.55495e-161 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15144, Q16728, Q8IUK3, Q8IVH3, Q9UCE0 | Gene names | ANPEP, APN, CD13, PEPN | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (hAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (gp150) (Myeloid plasma membrane glycoprotein CD13) (CD13 antigen). | |||||
|
AMPN_MOUSE
|
||||||
| NC score | 0.975583 (rank : 4) | θ value | 1.91815e-151 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97449, Q91YH8, Q99K63 | Gene names | Anpep, Lap-1, Lap1 | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (mAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (Membrane protein p161) (CD13 antigen). | |||||
|
ARTS1_MOUSE
|
||||||
| NC score | 0.974986 (rank : 5) | θ value | 3.49569e-153 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EQH2, Q9ET63 | Gene names | Arts1, Appils | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (VEGF-induced aminopeptidase). | |||||
|
ARTS1_HUMAN
|
||||||
| NC score | 0.974753 (rank : 6) | θ value | 6.38546e-147 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZ08, O60278, Q6UWY6, Q8NEL4, Q8TAD0, Q9UHF8, Q9UKY2 | Gene names | ARTS1, APPILS, KIAA0525 | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator). | |||||
|
LCAP_HUMAN
|
||||||
| NC score | 0.974423 (rank : 7) | θ value | 1.80368e-133 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIQ6, O00769, Q15145, Q9UIQ7 | Gene names | LNPEP, OTASE | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-cystinyl aminopeptidase (EC 3.4.11.3) (Cystinyl aminopeptidase) (Oxytocinase) (OTase) (Insulin-regulated membrane aminopeptidase) (Insulin-responsive aminopeptidase) (IRAP) (Placental leucine aminopeptidase) (P-LAP). | |||||
|
TRHDE_HUMAN
|
||||||
| NC score | 0.970483 (rank : 8) | θ value | 6.18479e-142 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKU6, Q6UWJ4 | Gene names | TRHDE | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
PSA_MOUSE
|
||||||
| NC score | 0.970038 (rank : 9) | θ value | 7.8238e-137 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
PSA_HUMAN
|
||||||
| NC score | 0.969990 (rank : 10) | θ value | 2.27638e-136 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
TRHDE_MOUSE
|
||||||
| NC score | 0.968984 (rank : 11) | θ value | 2.12569e-142 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K093 | Gene names | Trhde | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
LAEVR_HUMAN
|
||||||
| NC score | 0.963031 (rank : 12) | θ value | 2.14053e-118 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6Q4G3, Q32MR1, Q4G0I9, Q4G0V2, Q86XA3, Q8NBZ2 | Gene names | LVRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-) (CHL2 antigen). | |||||
|
LAEVR_MOUSE
|
||||||
| NC score | 0.955472 (rank : 13) | θ value | 7.70199e-76 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q2KHK3, Q9D633 | Gene names | Lvrn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-). | |||||
|
LKHA4_HUMAN
|
||||||
| NC score | 0.696002 (rank : 14) | θ value | 6.62687e-19 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09960 | Gene names | LTA4H, LTA4 | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
LKHA4_MOUSE
|
||||||
| NC score | 0.688492 (rank : 15) | θ value | 3.63628e-17 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24527, Q8VDR8 | Gene names | Lta4h | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
AMPB_MOUSE
|
||||||
| NC score | 0.651714 (rank : 16) | θ value | 3.40345e-15 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCT3 | Gene names | Rnpep | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase) (Cytosol aminopeptidase IV). | |||||
|
AMPB_HUMAN
|
||||||
| NC score | 0.636908 (rank : 17) | θ value | 2.43908e-13 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H4A4, Q9BVM9, Q9H1D4, Q9NPT7 | Gene names | RNPEP, APB | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase). | |||||
|
RNPL1_HUMAN
|
||||||
| NC score | 0.605235 (rank : 18) | θ value | 3.08544e-08 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
AMPO_HUMAN
|
||||||
| NC score | 0.444341 (rank : 19) | θ value | 1.29631e-06 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N6M6, Q5T9B1, Q5T9B3, Q5T9B4, Q8WUL6, Q96M23, Q96SS1 | Gene names | ONPEP, C9orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
AMPO_MOUSE
|
||||||
| NC score | 0.426821 (rank : 20) | θ value | 0.00390308 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BXQ6, Q6P0W6, Q6P394, Q8BHX5 | Gene names | Onpep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
TAF2_HUMAN
|
||||||
| NC score | 0.227822 (rank : 21) | θ value | 0.00228821 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1X5, O43487, O43604, O60668, Q86WW7, Q8IWK4 | Gene names | TAF2, CIF150, TAF2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa) (TAFII-150) (TAFII150) (150 kDa cofactor of initiator function). | |||||
|
TAF2_MOUSE
|
||||||
| NC score | 0.218881 (rank : 22) | θ value | 0.00298849 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C176, Q8BKQ7 | Gene names | Taf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa). | |||||
|
ARHG6_HUMAN
|
||||||
| NC score | 0.023933 (rank : 23) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
ARHG6_MOUSE
|
||||||
| NC score | 0.021507 (rank : 24) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K4I3, Q8C9V4 | Gene names | Arhgef6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6). | |||||
|
PACN2_HUMAN
|
||||||
| NC score | 0.020698 (rank : 25) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UNF0, O95921, Q96HV9, Q9H0D3, Q9NPN1, Q9Y4V2 | Gene names | PACSIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
ARHG7_HUMAN
|
||||||
| NC score | 0.020682 (rank : 26) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14155, Q6P9G3, Q6PII2, Q86W63, Q8N3M1 | Gene names | ARHGEF7, COOL1, KIAA0142, P85SPR, PAK3BP, PIXB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 7 (PAK-interacting exchange factor beta) (Beta-Pix) (COOL-1) (p85). | |||||
|
PACN2_MOUSE
|
||||||
| NC score | 0.020422 (rank : 27) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVE8 | Gene names | Pacsin2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
NF1_HUMAN
|
||||||
| NC score | 0.014488 (rank : 28) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21359, O00662, Q14284, Q14930, Q9UMK3 | Gene names | NF1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1) [Contains: Neurofibromin truncated]. | |||||
|
NF1_MOUSE
|
||||||
| NC score | 0.014443 (rank : 29) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04690, Q61956, Q61957 | Gene names | Nf1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofibromin (Neurofibromatosis-related protein NF-1). | |||||
|
RO60_MOUSE
|
||||||
| NC score | 0.012524 (rank : 30) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08848, Q9QYD8 | Gene names | Trove2, Ssa2 | |||
|
Domain Architecture |

|
|||||
| Description | 60 kDa SS-A/Ro ribonucleoprotein (60 kDa Ro protein) (60 kDa ribonucleoprotein Ro) (RoRNP) (TROVE domain family member 2). | |||||
|
RAD9A_MOUSE
|
||||||
| NC score | 0.011082 (rank : 31) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0F6, Q8QZZ3 | Gene names | Rad9a, Rad9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint control protein RAD9A (EC 3.1.11.2) (DNA repair exonuclease rad9 homolog A) (Rad9-like protein) (mRAD9). | |||||
|
OCTC_HUMAN
|
||||||
| NC score | 0.010583 (rank : 32) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKG9, Q8IUW9, Q9Y6I2 | Gene names | CROT, COT | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal carnitine O-octanoyltransferase (EC 2.3.1.137) (COT). | |||||
|
ECEL1_HUMAN
|
||||||
| NC score | 0.009876 (rank : 33) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95672, Q45UD9, Q53RF9, Q6UW86, Q86TH4, Q9NY95 | Gene names | ECEL1, XCE | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein). | |||||
|
ECEL1_MOUSE
|
||||||
| NC score | 0.009874 (rank : 34) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JMI0 | Gene names | Ecel1, Dine, Xce | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme-like 1 (EC 3.4.24.-) (Xce protein) (Damage-induced neuronal endopeptidase). | |||||
|
OBF1_MOUSE
|
||||||
| NC score | 0.009780 (rank : 35) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64693 | Gene names | Pou2af1, Obf-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain class 2-associating factor 1 (B-cell-specific coactivator OBF-1) (OCT-binding factor 1) (BOB-1) (BOB1) (OCA-B). | |||||
|
OCTC_MOUSE
|
||||||
| NC score | 0.007915 (rank : 36) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DC50, Q921I4 | Gene names | Crot, Cot | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal carnitine O-octanoyltransferase (EC 2.3.1.137) (COT). | |||||
|
TSNA1_HUMAN
|
||||||
| NC score | 0.005813 (rank : 37) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
ARFG1_MOUSE
|
||||||
| NC score | 0.005585 (rank : 38) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPJ9, Q3TI52, Q8BMM6, Q8BMQ7 | Gene names | Arfgap1, Arf1gap | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 1 (ADP-ribosylation factor 1 GTPase-activating protein) (ARF1 GAP) (ARF1-directed GTPase- activating protein) (GAP protein). | |||||
|
KLOT_MOUSE
|
||||||
| NC score | 0.005048 (rank : 39) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35082, O70175, O70621 | Gene names | Kl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Klotho precursor (EC 3.2.1.31) [Contains: Klotho peptide]. | |||||
|
ILEU_HUMAN
|
||||||
| NC score | 0.003796 (rank : 40) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30740, Q53FB9, Q5W0E1 | Gene names | SERPINB1, ELANH2, PI2 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte elastase inhibitor (LEI) (Serpin B1) (Monocyte/neutrophil elastase inhibitor) (M/NEI) (EI). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.003759 (rank : 41) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
HEP2_MOUSE
|
||||||
| NC score | 0.002746 (rank : 42) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49182 | Gene names | Serpind1, Hcf2, Hcii | |||
|
Domain Architecture |

|
|||||
| Description | Heparin cofactor 2 precursor (Heparin cofactor II) (HC-II) (Protease inhibitor leuserpin 2). | |||||
|
SYT12_HUMAN
|
||||||
| NC score | 0.002162 (rank : 43) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.002109 (rank : 44) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
DMD_MOUSE
|
||||||
| NC score | -0.000304 (rank : 45) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
CSKP_HUMAN
|
||||||
| NC score | -0.000421 (rank : 46) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||