Please be patient as the page loads
|
TRHDE_HUMAN
|
||||||
| SwissProt Accessions | Q9UKU6, Q6UWJ4 | Gene names | TRHDE | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRHDE_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKU6, Q6UWJ4 | Gene names | TRHDE | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
TRHDE_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998457 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K093 | Gene names | Trhde | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
AMPN_HUMAN
|
||||||
| θ value | 3.05528e-157 (rank : 3) | NC score | 0.975465 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15144, Q16728, Q8IUK3, Q8IVH3, Q9UCE0 | Gene names | ANPEP, APN, CD13, PEPN | |||
|
Domain Architecture |
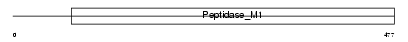
|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (hAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (gp150) (Myeloid plasma membrane glycoprotein CD13) (CD13 antigen). | |||||
|
AMPN_MOUSE
|
||||||
| θ value | 1.46868e-151 (rank : 4) | NC score | 0.974876 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97449, Q91YH8, Q99K63 | Gene names | Anpep, Lap-1, Lap1 | |||
|
Domain Architecture |
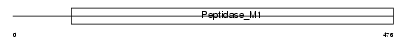
|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (mAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (Membrane protein p161) (CD13 antigen). | |||||
|
AMPE_HUMAN
|
||||||
| θ value | 6.18479e-142 (rank : 5) | NC score | 0.970483 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07075 | Gene names | ENPEP | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). | |||||
|
ARTS1_MOUSE
|
||||||
| θ value | 1.28961e-139 (rank : 6) | NC score | 0.968964 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EQH2, Q9ET63 | Gene names | Arts1, Appils | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (VEGF-induced aminopeptidase). | |||||
|
AMPE_MOUSE
|
||||||
| θ value | 2.87295e-139 (rank : 7) | NC score | 0.969839 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16406 | Gene names | Enpep | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (BP-1/6C3 antigen). | |||||
|
ARTS1_HUMAN
|
||||||
| θ value | 7.32286e-135 (rank : 8) | NC score | 0.968176 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZ08, O60278, Q6UWY6, Q8NEL4, Q8TAD0, Q9UHF8, Q9UKY2 | Gene names | ARTS1, APPILS, KIAA0525 | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator). | |||||
|
LAEVR_HUMAN
|
||||||
| θ value | 3.5201e-129 (rank : 9) | NC score | 0.965564 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6Q4G3, Q32MR1, Q4G0I9, Q4G0V2, Q86XA3, Q8NBZ2 | Gene names | LVRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-) (CHL2 antigen). | |||||
|
LCAP_HUMAN
|
||||||
| θ value | 2.61056e-124 (rank : 10) | NC score | 0.969045 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIQ6, O00769, Q15145, Q9UIQ7 | Gene names | LNPEP, OTASE | |||
|
Domain Architecture |
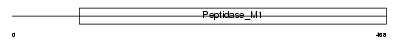
|
|||||
| Description | Leucyl-cystinyl aminopeptidase (EC 3.4.11.3) (Cystinyl aminopeptidase) (Oxytocinase) (OTase) (Insulin-regulated membrane aminopeptidase) (Insulin-responsive aminopeptidase) (IRAP) (Placental leucine aminopeptidase) (P-LAP). | |||||
|
PSA_HUMAN
|
||||||
| θ value | 1.87955e-106 (rank : 11) | NC score | 0.956285 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
PSA_MOUSE
|
||||||
| θ value | 2.71407e-105 (rank : 12) | NC score | 0.956134 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
LAEVR_MOUSE
|
||||||
| θ value | 3.12697e-77 (rank : 13) | NC score | 0.956629 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q2KHK3, Q9D633 | Gene names | Lvrn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-). | |||||
|
AMPB_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 14) | NC score | 0.660337 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCT3 | Gene names | Rnpep | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase) (Cytosol aminopeptidase IV). | |||||
|
LKHA4_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 15) | NC score | 0.694686 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09960 | Gene names | LTA4H, LTA4 | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
LKHA4_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 16) | NC score | 0.687743 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24527, Q8VDR8 | Gene names | Lta4h | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
AMPB_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 17) | NC score | 0.646686 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H4A4, Q9BVM9, Q9H1D4, Q9NPT7 | Gene names | RNPEP, APB | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase). | |||||
|
RNPL1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 18) | NC score | 0.621329 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
AMPO_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 19) | NC score | 0.464158 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N6M6, Q5T9B1, Q5T9B3, Q5T9B4, Q8WUL6, Q96M23, Q96SS1 | Gene names | ONPEP, C9orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
AMPO_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 20) | NC score | 0.451339 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BXQ6, Q6P0W6, Q6P394, Q8BHX5 | Gene names | Onpep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
TAF2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.199704 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1X5, O43487, O43604, O60668, Q86WW7, Q8IWK4 | Gene names | TAF2, CIF150, TAF2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa) (TAFII-150) (TAFII150) (150 kDa cofactor of initiator function). | |||||
|
EDAD_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.056175 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWZ3, Q5VYJ7 | Gene names | EDARADD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectodysplasin A receptor-associated adapter protein (EDAR-associated death domain protein) (Crinkled homolog). | |||||
|
TAF2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.190569 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C176, Q8BKQ7 | Gene names | Taf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.025819 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.025745 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO5A1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.023145 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO5A1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.023186 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
ROA3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.008284 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51991, Q6URK5 | Gene names | HNRPA3 | |||
|
Domain Architecture |
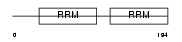
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
ROA3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.008247 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |
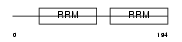
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
PRIO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.018322 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04156, O60489, P78446, Q15216, Q15221, Q8TBG0, Q96E70, Q9UP19 | Gene names | PRNP, PRP | |||
|
Domain Architecture |
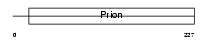
|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (ASCR) (CD230 antigen). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.016872 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
CO1A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.021770 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.016624 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.018791 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.016339 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
FBN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.000705 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
CITE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.014740 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99967, O95426 | Gene names | CITED2, MRG1 | |||
|
Domain Architecture |
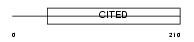
|
|||||
| Description | Cbp/p300-interacting transactivator 2 (MSG-related protein 1) (MRG-1) (P35srj). | |||||
|
CO2A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.017921 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |
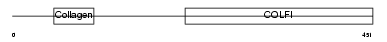
|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.013433 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
PSG9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.004906 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00887, Q15236, Q15237, Q8WW78, Q9UQ73 | Gene names | PSG9, PSG11 | |||
|
Domain Architecture |
|
|||||
| Description | Pregnancy-specific beta-1-glycoprotein 9 precursor (PSBG-9) (Pregnancy-specific glycoprotein 9) (Pregnancy-specific beta-1 glycoprotein B) (PS-beta-B) (PS34) (Pregnancy-specific glycoprotein 7) (PSG7). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.003912 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.014205 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TPSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.009595 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R233, O70317, Q91YE0, Q91YE1, Q9QWT7, Q9Z1W3 | Gene names | Tapbp, Tapa | |||
|
Domain Architecture |

|
|||||
| Description | Tapasin precursor (TPSN) (TPN) (TAP-binding protein) (TAP-associated protein). | |||||
|
CITE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.012469 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35740, O35741, O35742, O35743, O55198 | Gene names | Cited2, Mrg1, Msg2 | |||
|
Domain Architecture |
|
|||||
| Description | Cbp/p300-interacting transactivator 2 (MSG-related protein 1) (MRG-1). | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.016032 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.014338 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | -0.002400 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
NPT2C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.004951 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N130 | Gene names | SLC34A3, NPT2C, NPTIIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-dependent phosphate transport protein 2C (Sodium/phosphate cotransporter 2C) (Na(+)/Pi cotransporter 2C) (Sodium-phosphate transport protein 2C) (Na(+)-dependent phosphate cotransporter 2C) (NaPi-2c) (Sodium/inorganic phosphate cotransporter IIC) (Solute carrier family 34 member 3). | |||||
|
TAF5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | -0.000146 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C092, Q8BTR2 | Gene names | Taf5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
TRHDE_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UKU6, Q6UWJ4 | Gene names | TRHDE | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
TRHDE_MOUSE
|
||||||
| NC score | 0.998457 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K093 | Gene names | Trhde | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotropin-releasing hormone-degrading ectoenzyme (EC 3.4.19.6) (TRH- degrading ectoenzyme) (TRH-DE) (TRH-specific aminopeptidase) (Thyroliberinase) (Pyroglutamyl-peptidase II) (PAP-II). | |||||
|
AMPN_HUMAN
|
||||||
| NC score | 0.975465 (rank : 3) | θ value | 3.05528e-157 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15144, Q16728, Q8IUK3, Q8IVH3, Q9UCE0 | Gene names | ANPEP, APN, CD13, PEPN | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (hAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (gp150) (Myeloid plasma membrane glycoprotein CD13) (CD13 antigen). | |||||
|
AMPN_MOUSE
|
||||||
| NC score | 0.974876 (rank : 4) | θ value | 1.46868e-151 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97449, Q91YH8, Q99K63 | Gene names | Anpep, Lap-1, Lap1 | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase N (EC 3.4.11.2) (mAPN) (Alanyl aminopeptidase) (Microsomal aminopeptidase) (Aminopeptidase M) (Membrane protein p161) (CD13 antigen). | |||||
|
AMPE_HUMAN
|
||||||
| NC score | 0.970483 (rank : 5) | θ value | 6.18479e-142 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07075 | Gene names | ENPEP | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). | |||||
|
AMPE_MOUSE
|
||||||
| NC score | 0.969839 (rank : 6) | θ value | 2.87295e-139 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16406 | Gene names | Enpep | |||
|
Domain Architecture |

|
|||||
| Description | Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (BP-1/6C3 antigen). | |||||
|
LCAP_HUMAN
|
||||||
| NC score | 0.969045 (rank : 7) | θ value | 2.61056e-124 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIQ6, O00769, Q15145, Q9UIQ7 | Gene names | LNPEP, OTASE | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-cystinyl aminopeptidase (EC 3.4.11.3) (Cystinyl aminopeptidase) (Oxytocinase) (OTase) (Insulin-regulated membrane aminopeptidase) (Insulin-responsive aminopeptidase) (IRAP) (Placental leucine aminopeptidase) (P-LAP). | |||||
|
ARTS1_MOUSE
|
||||||
| NC score | 0.968964 (rank : 8) | θ value | 1.28961e-139 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EQH2, Q9ET63 | Gene names | Arts1, Appils | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (VEGF-induced aminopeptidase). | |||||
|
ARTS1_HUMAN
|
||||||
| NC score | 0.968176 (rank : 9) | θ value | 7.32286e-135 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZ08, O60278, Q6UWY6, Q8NEL4, Q8TAD0, Q9UHF8, Q9UKY2 | Gene names | ARTS1, APPILS, KIAA0525 | |||
|
Domain Architecture |

|
|||||
| Description | Adipocyte-derived leucine aminopeptidase precursor (EC 3.4.11.-) (A- LAP) (ARTS-1) (Aminopeptidase PILS) (Puromycin-insensitive leucyl- specific aminopeptidase) (PILS-AP) (Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator). | |||||
|
LAEVR_HUMAN
|
||||||
| NC score | 0.965564 (rank : 10) | θ value | 3.5201e-129 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6Q4G3, Q32MR1, Q4G0I9, Q4G0V2, Q86XA3, Q8NBZ2 | Gene names | LVRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-) (CHL2 antigen). | |||||
|
LAEVR_MOUSE
|
||||||
| NC score | 0.956629 (rank : 11) | θ value | 3.12697e-77 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q2KHK3, Q9D633 | Gene names | Lvrn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-). | |||||
|
PSA_HUMAN
|
||||||
| NC score | 0.956285 (rank : 12) | θ value | 1.87955e-106 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
PSA_MOUSE
|
||||||
| NC score | 0.956134 (rank : 13) | θ value | 2.71407e-105 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
LKHA4_HUMAN
|
||||||
| NC score | 0.694686 (rank : 14) | θ value | 1.80466e-16 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P09960 | Gene names | LTA4H, LTA4 | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
LKHA4_MOUSE
|
||||||
| NC score | 0.687743 (rank : 15) | θ value | 3.07829e-16 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24527, Q8VDR8 | Gene names | Lta4h | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene A-4 hydrolase (EC 3.3.2.6) (LTA-4 hydrolase) (Leukotriene A(4) hydrolase). | |||||
|
AMPB_MOUSE
|
||||||
| NC score | 0.660337 (rank : 16) | θ value | 1.058e-16 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCT3 | Gene names | Rnpep | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase) (Cytosol aminopeptidase IV). | |||||
|
AMPB_HUMAN
|
||||||
| NC score | 0.646686 (rank : 17) | θ value | 7.58209e-15 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H4A4, Q9BVM9, Q9H1D4, Q9NPT7 | Gene names | RNPEP, APB | |||
|
Domain Architecture |

|
|||||
| Description | Aminopeptidase B (EC 3.4.11.6) (Ap-B) (Arginyl aminopeptidase) (Arginine aminopeptidase). | |||||
|
RNPL1_HUMAN
|
||||||
| NC score | 0.621329 (rank : 18) | θ value | 6.64225e-11 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU8, Q96AC9, Q9H033, Q9NVD0 | Gene names | RNPEPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl aminopeptidase-like 1 (EC 3.4.11.-) (RNPEP-like protein). | |||||
|
AMPO_HUMAN
|
||||||
| NC score | 0.464158 (rank : 19) | θ value | 4.45536e-07 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N6M6, Q5T9B1, Q5T9B3, Q5T9B4, Q8WUL6, Q96M23, Q96SS1 | Gene names | ONPEP, C9orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
AMPO_MOUSE
|
||||||
| NC score | 0.451339 (rank : 20) | θ value | 7.59969e-07 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BXQ6, Q6P0W6, Q6P394, Q8BHX5 | Gene names | Onpep | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aminopeptidase O (EC 3.4.11.-) (AP-O). | |||||
|
TAF2_HUMAN
|
||||||
| NC score | 0.199704 (rank : 21) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1X5, O43487, O43604, O60668, Q86WW7, Q8IWK4 | Gene names | TAF2, CIF150, TAF2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa) (TAFII-150) (TAFII150) (150 kDa cofactor of initiator function). | |||||
|
TAF2_MOUSE
|
||||||
| NC score | 0.190569 (rank : 22) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C176, Q8BKQ7 | Gene names | Taf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 2 (Transcription initiation factor TFIID 150 kDa subunit) (TBP-associated factor 150 kDa). | |||||
|
EDAD_HUMAN
|
||||||
| NC score | 0.056175 (rank : 23) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WWZ3, Q5VYJ7 | Gene names | EDARADD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectodysplasin A receptor-associated adapter protein (EDAR-associated death domain protein) (Crinkled homolog). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.025819 (rank : 24) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.025745 (rank : 25) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO5A1_MOUSE
|
||||||
| NC score | 0.023186 (rank : 26) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO5A1_HUMAN
|
||||||
| NC score | 0.023145 (rank : 27) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO1A1_MOUSE
|
||||||
| NC score | 0.021770 (rank : 28) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.018791 (rank : 29) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
PRIO_HUMAN
|
||||||
| NC score | 0.018322 (rank : 30) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04156, O60489, P78446, Q15216, Q15221, Q8TBG0, Q96E70, Q9UP19 | Gene names | PRNP, PRP | |||
|
Domain Architecture |

|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (ASCR) (CD230 antigen). | |||||
|
CO2A1_HUMAN
|
||||||
| NC score | 0.017921 (rank : 31) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.016872 (rank : 32) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.016624 (rank : 33) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.016339 (rank : 34) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.016032 (rank : 35) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CITE2_HUMAN
|
||||||
| NC score | 0.014740 (rank : 36) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99967, O95426 | Gene names | CITED2, MRG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 2 (MSG-related protein 1) (MRG-1) (P35srj). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.014338 (rank : 37) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.014205 (rank : 38) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.013433 (rank : 39) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
CITE2_MOUSE
|
||||||
| NC score | 0.012469 (rank : 40) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35740, O35741, O35742, O35743, O55198 | Gene names | Cited2, Mrg1, Msg2 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 2 (MSG-related protein 1) (MRG-1). | |||||
|
TPSN_MOUSE
|
||||||
| NC score | 0.009595 (rank : 41) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R233, O70317, Q91YE0, Q91YE1, Q9QWT7, Q9Z1W3 | Gene names | Tapbp, Tapa | |||
|
Domain Architecture |

|
|||||
| Description | Tapasin precursor (TPSN) (TPN) (TAP-binding protein) (TAP-associated protein). | |||||
|
ROA3_HUMAN
|
||||||
| NC score | 0.008284 (rank : 42) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51991, Q6URK5 | Gene names | HNRPA3 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
ROA3_MOUSE
|
||||||
| NC score | 0.008247 (rank : 43) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG05, Q8BHF8 | Gene names | Hnrpa3, Hnrnpa3 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein A3 (hnRNP A3). | |||||
|
NPT2C_HUMAN
|
||||||
| NC score | 0.004951 (rank : 44) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N130 | Gene names | SLC34A3, NPT2C, NPTIIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-dependent phosphate transport protein 2C (Sodium/phosphate cotransporter 2C) (Na(+)/Pi cotransporter 2C) (Sodium-phosphate transport protein 2C) (Na(+)-dependent phosphate cotransporter 2C) (NaPi-2c) (Sodium/inorganic phosphate cotransporter IIC) (Solute carrier family 34 member 3). | |||||
|
PSG9_HUMAN
|
||||||
| NC score | 0.004906 (rank : 45) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00887, Q15236, Q15237, Q8WW78, Q9UQ73 | Gene names | PSG9, PSG11 | |||
|
Domain Architecture |

|
|||||
| Description | Pregnancy-specific beta-1-glycoprotein 9 precursor (PSBG-9) (Pregnancy-specific glycoprotein 9) (Pregnancy-specific beta-1 glycoprotein B) (PS-beta-B) (PS34) (Pregnancy-specific glycoprotein 7) (PSG7). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.003912 (rank : 46) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.000705 (rank : 47) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
TAF5_MOUSE
|
||||||
| NC score | -0.000146 (rank : 48) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C092, Q8BTR2 | Gene names | Taf5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | -0.002400 (rank : 49) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||