Please be patient as the page loads
|
PKP4_HUMAN
|
||||||
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CTND2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.951556 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.964321 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
PKP4_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993729 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
ARVC_HUMAN
|
||||||
| θ value | 2.97994e-128 (rank : 5) | NC score | 0.945767 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
ARVC_MOUSE
|
||||||
| θ value | 3.64273e-126 (rank : 6) | NC score | 0.947461 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
CTND1_MOUSE
|
||||||
| θ value | 1.58377e-121 (rank : 7) | NC score | 0.942942 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_HUMAN
|
||||||
| θ value | 1.93603e-119 (rank : 8) | NC score | 0.940777 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
PKP1_MOUSE
|
||||||
| θ value | 3.34864e-71 (rank : 9) | NC score | 0.902917 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
PKP1_HUMAN
|
||||||
| θ value | 9.11921e-69 (rank : 10) | NC score | 0.901647 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
PKP2_HUMAN
|
||||||
| θ value | 7.71989e-68 (rank : 11) | NC score | 0.909864 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
PKP3_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 12) | NC score | 0.898959 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP3_MOUSE
|
||||||
| θ value | 1.50663e-63 (rank : 13) | NC score | 0.898165 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
APC_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 14) | NC score | 0.236845 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
APC_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 15) | NC score | 0.231467 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
IMA7_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 16) | NC score | 0.187907 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |
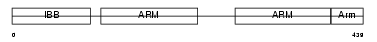
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
IMA7_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 17) | NC score | 0.183361 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |
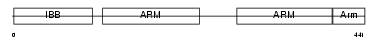
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
IMA2_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 18) | NC score | 0.168865 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |
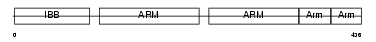
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 19) | NC score | 0.169775 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |
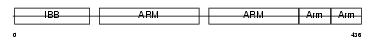
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 20) | NC score | 0.365895 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
CTNB1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 21) | NC score | 0.272055 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 22) | NC score | 0.272120 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
IMA4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 23) | NC score | 0.161982 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |
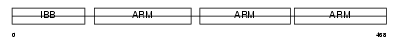
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 24) | NC score | 0.161978 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |
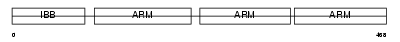
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
PLAK_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 25) | NC score | 0.204873 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
IMA5_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 26) | NC score | 0.175243 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
HIF1N_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.054908 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLR9, Q3U3G4 | Gene names | Hif1an | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha inhibitor (EC 1.14.11.16) (Hypoxia- inducible factor asparagine hydroxylase). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.050494 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.016919 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
IMA1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.147843 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
IMA1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.147584 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
IMA3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.154532 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |
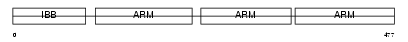
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.154663 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |
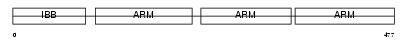
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
PLAK_MOUSE
|
||||||
| θ value | 0.163984 (rank : 34) | NC score | 0.241204 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.014882 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 36) | NC score | 0.050707 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.035747 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
CCD27_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.023457 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
EPCR_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.049014 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNN8, Q14218, Q6IB56, Q96CB3, Q9ULX1 | Gene names | PROCR, EPCR | |||
|
Domain Architecture |
|
|||||
| Description | Endothelial protein C receptor precursor (Endothelial cell protein C receptor) (Activated protein C receptor) (APC receptor) (CD201 antigen). | |||||
|
PRC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.032505 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43663, Q9BSB6 | Gene names | PRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.022977 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.040775 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.020168 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.032841 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
HEAT3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.079105 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
WT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.001889 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P19544, Q15881, Q16256, Q16575, Q8IYZ5 | Gene names | WT1 | |||
|
Domain Architecture |

|
|||||
| Description | Wilms' tumor protein (WT33). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.027494 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.012463 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MTMR7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.016259 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
PCDA9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.002619 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.022911 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.041629 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DAG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.034721 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14118 | Gene names | DAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
GDS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.174868 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.020568 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.024462 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.031322 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.004602 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
COE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.020905 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.020905 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.001187 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.024092 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.010309 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.003406 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
DAG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.028692 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.041385 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
GAB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.022444 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
HIF1N_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.035363 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWT6, Q969Q7, Q9NPV5 | Gene names | HIF1AN, FIH1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha inhibitor (EC 1.14.11.16) (Hypoxia- inducible factor asparagine hydroxylase) (Factor inhibiting HIF-1) (FIH-1). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.045116 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.016671 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ZBTB3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.003374 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
MUC13_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.015544 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3R2, Q6UWD9, Q9NXT5 | Gene names | MUC13, DRCC1, RECC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-13 precursor (Down-regulated in colon cancer 1). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.004936 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
RTDR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.058602 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
SMN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.041402 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |
|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
TLN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.011188 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
WDR79_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.009563 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BUR4, Q9NW09 | Gene names | WDR79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 79. | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.006168 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.017739 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
K1718_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.019856 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.005518 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.015677 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.015203 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
WT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | -0.000023 (rank : 110) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22561 | Gene names | Wt1, Wt-1 | |||
|
Domain Architecture |
|
|||||
| Description | Wilms' tumor protein homolog. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.058592 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BAP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.020699 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
P66A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.015232 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.025365 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.002681 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.000513 (rank : 109) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.019268 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.019443 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.004411 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.018819 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
GP125_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.003047 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWK6, Q6UXK9, Q86SQ5, Q8TC55 | Gene names | GPR125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
HEAT3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.051945 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |
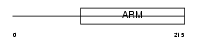
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
KCNN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.004686 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.013969 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.003242 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
P66A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.012601 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.000537 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ABL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | -0.001882 (rank : 111) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.004935 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
FOXE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.001026 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00358, O75765 | Gene names | FOXE1, FKHL15, TITF2, TTF2 | |||
|
Domain Architecture |
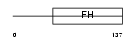
|
|||||
| Description | Forkhead box protein E1 (Thyroid transcription factor 2) (TTF-2) (Forkhead-related protein FKHL15). | |||||
|
MTMR7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.008830 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.049027 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZAP70_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | -0.002602 (rank : 112) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43403 | Gene names | ZAP70, SRK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ZAP-70 (EC 2.7.10.2) (70 kDa zeta-associated protein) (Syk-related tyrosine kinase). | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.055352 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.057270 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.054049 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.068854 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
SPAG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.061694 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
PKP4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.993729 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.964321 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.951556 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
ARVC_MOUSE
|
||||||
| NC score | 0.947461 (rank : 5) | θ value | 3.64273e-126 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.945767 (rank : 6) | θ value | 2.97994e-128 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
CTND1_MOUSE
|
||||||
| NC score | 0.942942 (rank : 7) | θ value | 1.58377e-121 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.940777 (rank : 8) | θ value | 1.93603e-119 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
PKP2_HUMAN
|
||||||
| NC score | 0.909864 (rank : 9) | θ value | 7.71989e-68 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
PKP1_MOUSE
|
||||||
| NC score | 0.902917 (rank : 10) | θ value | 3.34864e-71 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
PKP1_HUMAN
|
||||||
| NC score | 0.901647 (rank : 11) | θ value | 9.11921e-69 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
PKP3_HUMAN
|
||||||
| NC score | 0.898959 (rank : 12) | θ value | 1.50663e-63 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP3_MOUSE
|
||||||
| NC score | 0.898165 (rank : 13) | θ value | 1.50663e-63 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.365895 (rank : 14) | θ value | 0.00298849 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
CTNB1_MOUSE
|
||||||
| NC score | 0.272120 (rank : 15) | θ value | 0.00509761 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_HUMAN
|
||||||
| NC score | 0.272055 (rank : 16) | θ value | 0.00509761 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
PLAK_MOUSE
|
||||||
| NC score | 0.241204 (rank : 17) | θ value | 0.163984 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.236845 (rank : 18) | θ value | 4.1701e-05 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.231467 (rank : 19) | θ value | 5.44631e-05 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
PLAK_HUMAN
|
||||||
| NC score | 0.204873 (rank : 20) | θ value | 0.00665767 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
IMA7_MOUSE
|
||||||
| NC score | 0.187907 (rank : 21) | θ value | 0.000121331 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
IMA7_HUMAN
|
||||||
| NC score | 0.183361 (rank : 22) | θ value | 0.00035302 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
IMA5_HUMAN
|
||||||
| NC score | 0.175243 (rank : 23) | θ value | 0.0148317 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
GDS1_HUMAN
|
||||||
| NC score | 0.174868 (rank : 24) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
IMA2_MOUSE
|
||||||
| NC score | 0.169775 (rank : 25) | θ value | 0.00175202 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA2_HUMAN
|
||||||
| NC score | 0.168865 (rank : 26) | θ value | 0.00175202 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA4_HUMAN
|
||||||
| NC score | 0.161982 (rank : 27) | θ value | 0.00509761 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| NC score | 0.161978 (rank : 28) | θ value | 0.00509761 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
IMA3_MOUSE
|
||||||
| NC score | 0.154663 (rank : 29) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
IMA3_HUMAN
|
||||||
| NC score | 0.154532 (rank : 30) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA1_HUMAN
|
||||||
| NC score | 0.147843 (rank : 31) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
IMA1_MOUSE
|
||||||
| NC score | 0.147584 (rank : 32) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
HEAT3_MOUSE
|
||||||
| NC score | 0.079105 (rank : 33) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.068854 (rank : 34) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
SPAG6_MOUSE
|
||||||
| NC score | 0.061694 (rank : 35) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
RTDR1_HUMAN
|
||||||
| NC score | 0.058602 (rank : 36) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.058592 (rank : 37) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.057270 (rank : 38) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.055352 (rank : 39) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
HIF1N_MOUSE
|
||||||
| NC score | 0.054908 (rank : 40) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLR9, Q3U3G4 | Gene names | Hif1an | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha inhibitor (EC 1.14.11.16) (Hypoxia- inducible factor asparagine hydroxylase). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.054049 (rank : 41) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
HEAT3_HUMAN
|
||||||
| NC score | 0.051945 (rank : 42) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.050707 (rank : 43) | θ value | 0.21417 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.050494 (rank : 44) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.049027 (rank : 45) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
EPCR_HUMAN
|
||||||
| NC score | 0.049014 (rank : 46) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNN8, Q14218, Q6IB56, Q96CB3, Q9ULX1 | Gene names | PROCR, EPCR | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial protein C receptor precursor (Endothelial cell protein C receptor) (Activated protein C receptor) (APC receptor) (CD201 antigen). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.045116 (rank : 47) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.041629 (rank : 48) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SMN_HUMAN
|
||||||
| NC score | 0.041402 (rank : 49) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |

|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.041385 (rank : 50) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.040775 (rank : 51) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.035747 (rank : 52) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
HIF1N_HUMAN
|
||||||
| NC score | 0.035363 (rank : 53) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWT6, Q969Q7, Q9NPV5 | Gene names | HIF1AN, FIH1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha inhibitor (EC 1.14.11.16) (Hypoxia- inducible factor asparagine hydroxylase) (Factor inhibiting HIF-1) (FIH-1). | |||||
|
DAG1_HUMAN
|
||||||
| NC score | 0.034721 (rank : 54) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14118 | Gene names | DAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.032841 (rank : 55) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
PRC1_HUMAN
|
||||||
| NC score | 0.032505 (rank : 56) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43663, Q9BSB6 | Gene names | PRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.031322 (rank : 57) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
DAG1_MOUSE
|
||||||
| NC score | 0.028692 (rank : 58) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62165, Q61094, Q61141, Q61497 | Gene names | Dag1, Dag-1 | |||
|
Domain Architecture |

|
|||||
| Description | Dystroglycan precursor (Dystrophin-associated glycoprotein 1) [Contains: Alpha-dystroglycan (Alpha-DG); Beta-dystroglycan (Beta- DG)]. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.027494 (rank : 59) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.025365 (rank : 60) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.024462 (rank : 61) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.024092 (rank : 62) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CCD27_MOUSE
|
||||||
| NC score | 0.023457 (rank : 63) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.022977 (rank : 64) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.022911 (rank : 65) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.022444 (rank : 66) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
COE1_HUMAN
|
||||||
| NC score | 0.020905 (rank : 67) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| NC score | 0.020905 (rank : 68) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
BAP1_HUMAN
|
||||||
| NC score | 0.020699 (rank : 69) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92560, Q6LEM0, Q7Z5E8 | Gene names | BAP1, KIAA0272 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase BAP1 (EC 3.4.19.12) (BRCA1- associated protein 1) (Cerebral protein 6). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.020568 (rank : 70) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.020168 (rank : 71) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
K1718_HUMAN
|
||||||
| NC score | 0.019856 (rank : 72) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.019443 (rank : 73) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.019268 (rank : 74) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.018819 (rank : 75) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.017739 (rank : 76) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.016919 (rank : 77) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.016671 (rank : 78) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
MTMR7_HUMAN
|
||||||
| NC score | 0.016259 (rank : 79) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.015677 (rank : 80) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
MUC13_HUMAN
|
||||||
| NC score | 0.015544 (rank : 81) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3R2, Q6UWD9, Q9NXT5 | Gene names | MUC13, DRCC1, RECC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-13 precursor (Down-regulated in colon cancer 1). | |||||
|
P66A_HUMAN
|
||||||
| NC score | 0.015232 (rank : 82) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YP4, Q7L3J2, Q96F28, Q9NPU2, Q9NXS1 | Gene names | GATAD2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (Hp66alpha) (GATA zinc finger domain-containing protein 2A). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.015203 (rank : 83) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.014882 (rank : 84) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.013969 (rank : 85) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
P66A_MOUSE
|
||||||
| NC score | 0.012601 (rank : 86) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.012463 (rank : 87) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TLN2_MOUSE
|
||||||
| NC score | 0.011188 (rank : 88) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.010309 (rank : 89) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
WDR79_HUMAN
|
||||||
| NC score | 0.009563 (rank : 90) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BUR4, Q9NW09 | Gene names | WDR79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 79. | |||||
|
MTMR7_MOUSE
|
||||||
| NC score | 0.008830 (rank : 91) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.006168 (rank : 92) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.005518 (rank : 93) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.004936 (rank : 94) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.004935 (rank : 95) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
KCNN1_MOUSE
|
||||||
| NC score | 0.004686 (rank : 96) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.004602 (rank : 97) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.004411 (rank : 98) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | 0.003406 (rank : 99) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
ZBTB3_HUMAN
|
||||||
| NC score | 0.003374 (rank : 100) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H5J0 | Gene names | ZBTB3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 3. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.003242 (rank : 101) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GP125_HUMAN
|
||||||
| NC score | 0.003047 (rank : 102) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWK6, Q6UXK9, Q86SQ5, Q8TC55 | Gene names | GPR125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.002681 (rank : 103) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
PCDA9_HUMAN
|
||||||
| NC score | 0.002619 (rank : 104) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
WT1_HUMAN
|
||||||
| NC score | 0.001889 (rank : 105) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P19544, Q15881, Q16256, Q16575, Q8IYZ5 | Gene names | WT1 | |||
|
Domain Architecture |

|
|||||
| Description | Wilms' tumor protein (WT33). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.001187 (rank : 106) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
FOXE1_HUMAN
|
||||||
| NC score | 0.001026 (rank : 107) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00358, O75765 | Gene names | FOXE1, FKHL15, TITF2, TTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E1 (Thyroid transcription factor 2) (TTF-2) (Forkhead-related protein FKHL15). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.000537 (rank : 108) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.000513 (rank : 109) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
WT1_MOUSE
|
||||||
| NC score | -0.000023 (rank : 110) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22561 | Gene names | Wt1, Wt-1 | |||
|
Domain Architecture |

|
|||||
| Description | Wilms' tumor protein homolog. | |||||
|
ABL2_HUMAN
|
||||||
| NC score | -0.001882 (rank : 111) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
ZAP70_HUMAN
|
||||||
| NC score | -0.002602 (rank : 112) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43403 | Gene names | ZAP70, SRK | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ZAP-70 (EC 2.7.10.2) (70 kDa zeta-associated protein) (Syk-related tyrosine kinase). | |||||