Please be patient as the page loads
|
MTMR7_HUMAN
|
||||||
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MTMR6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988440 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y217, Q86TB7, Q86YH4, Q96P80 | Gene names | MTMR6 | |||
|
Domain Architecture |
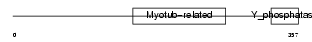
|
|||||
| Description | Myotubularin-related protein 6 (EC 3.1.3.-). | |||||
|
MTMR7_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR7_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.997099 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR2_HUMAN
|
||||||
| θ value | 4.19693e-98 (rank : 4) | NC score | 0.952476 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13614, Q9UPS9 | Gene names | MTMR2, KIAA1073 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTMR2_MOUSE
|
||||||
| θ value | 7.91507e-97 (rank : 5) | NC score | 0.952331 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D1, Q8VHA7 | Gene names | Mtmr2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTM1_HUMAN
|
||||||
| θ value | 2.15548e-94 (rank : 6) | NC score | 0.951871 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13496, Q8NEL1 | Gene names | MTM1, CG2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTMR9_HUMAN
|
||||||
| θ value | 6.27148e-94 (rank : 7) | NC score | 0.960857 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QG7, Q52LU3, Q8WW11, Q96QG6, Q9NX50 | Gene names | MTMR9, C8orf9, MTMR8 | |||
|
Domain Architecture |
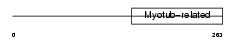
|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR9_MOUSE
|
||||||
| θ value | 6.27148e-94 (rank : 8) | NC score | 0.961141 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D0, Q80XL4 | Gene names | Mtmr9, Mtmr3 | |||
|
Domain Architecture |
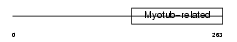
|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTM1_MOUSE
|
||||||
| θ value | 2.38316e-93 (rank : 9) | NC score | 0.951452 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2C5 | Gene names | Mtm1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTMR1_MOUSE
|
||||||
| θ value | 9.37149e-90 (rank : 10) | NC score | 0.949733 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2C4 | Gene names | Mtmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR1_HUMAN
|
||||||
| θ value | 6.07444e-89 (rank : 11) | NC score | 0.949196 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13613, Q9UBX6, Q9UEM0, Q9UQD5 | Gene names | MTMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR3_HUMAN
|
||||||
| θ value | 1.71583e-75 (rank : 12) | NC score | 0.824098 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
MTMR3_MOUSE
|
||||||
| θ value | 2.92676e-75 (rank : 13) | NC score | 0.925299 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |
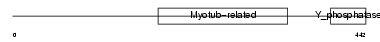
|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 14) | NC score | 0.698153 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
MTMRD_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 15) | NC score | 0.692878 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.050223 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.052371 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
PCTK2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.006718 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0D0 | Gene names | Pctk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PCTAIRE-2 (EC 2.7.11.22) (PCTAIRE- motif protein kinase 2). | |||||
|
LYRIC_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.063181 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.011135 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
PCTK2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.006181 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00537, Q8NEB8 | Gene names | PCTK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PCTAIRE-2 (EC 2.7.11.22) (PCTAIRE- motif protein kinase 2). | |||||
|
JHD2A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.027368 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
YETS2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.028512 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
DYH9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.020119 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.028791 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.024578 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.030276 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
LYRIC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.051575 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
PKP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.016259 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.010362 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
ELOA3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.016917 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
HNRPC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.021801 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |
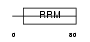
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.003251 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.018723 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
DMRTA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.017691 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFG4 | Gene names | Dmrta1, Dmrt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor A1 (Doublesex- and mab-3-related transcription factor 4). | |||||
|
DUS5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.016574 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
HPS5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.024078 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPZ3, O95942, Q8N4U0 | Gene names | HPS5, AIBP63, KIAA1017 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein (Alpha-integrin-binding protein 63) (Ruby-eye protein 2 homolog) (Ru2). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.019966 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.003160 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
AZI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.007140 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
LIGA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.025648 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61211, Q3TDE0, Q3THV5, Q3TTP4, Q8C491, Q8CBF1, Q8CC17, Q8R1I9, Q8R3M5 | Gene names | Lgtn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligatin. | |||||
|
NEMO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.008707 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.008884 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.015199 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.010271 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
IPP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.016542 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.010547 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
AMBN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.012036 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP70, Q9H2X1, Q9H4L1 | Gene names | AMBN | |||
|
Domain Architecture |
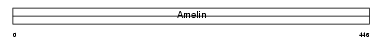
|
|||||
| Description | Ameloblastin precursor. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.008704 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
HOME1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.013393 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |
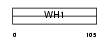
|
|||||
| Description | Homer protein homolog 1. | |||||
|
HOME1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.012740 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.012309 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.008679 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SPC24_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.012755 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D083, Q3TIF5, Q8CD13 | Gene names | Spbc24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24. | |||||
|
TACC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.014063 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.021409 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.036770 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.013692 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.004084 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.011138 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
DUS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.012781 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.014342 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.003479 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.013616 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CD45_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.004553 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.004065 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.007225 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
K1024_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.012466 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPX6 | Gene names | KIAA1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024. | |||||
|
PFD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.015874 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQP4, Q5TD11, Q92779 | Gene names | PFDN4, PFD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prefoldin subunit 4 (Protein C-1). | |||||
|
SG16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.046246 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
UTY_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.005440 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
DEN4C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.050774 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
MYCPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.061422 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051537 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.052357 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
MTMR7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR7_MOUSE
|
||||||
| NC score | 0.997099 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR6_HUMAN
|
||||||
| NC score | 0.988440 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y217, Q86TB7, Q86YH4, Q96P80 | Gene names | MTMR6 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 6 (EC 3.1.3.-). | |||||
|
MTMR9_MOUSE
|
||||||
| NC score | 0.961141 (rank : 4) | θ value | 6.27148e-94 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D0, Q80XL4 | Gene names | Mtmr9, Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR9_HUMAN
|
||||||
| NC score | 0.960857 (rank : 5) | θ value | 6.27148e-94 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QG7, Q52LU3, Q8WW11, Q96QG6, Q9NX50 | Gene names | MTMR9, C8orf9, MTMR8 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR2_HUMAN
|
||||||
| NC score | 0.952476 (rank : 6) | θ value | 4.19693e-98 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13614, Q9UPS9 | Gene names | MTMR2, KIAA1073 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTMR2_MOUSE
|
||||||
| NC score | 0.952331 (rank : 7) | θ value | 7.91507e-97 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D1, Q8VHA7 | Gene names | Mtmr2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTM1_HUMAN
|
||||||
| NC score | 0.951871 (rank : 8) | θ value | 2.15548e-94 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13496, Q8NEL1 | Gene names | MTM1, CG2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTM1_MOUSE
|
||||||
| NC score | 0.951452 (rank : 9) | θ value | 2.38316e-93 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2C5 | Gene names | Mtm1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTMR1_MOUSE
|
||||||
| NC score | 0.949733 (rank : 10) | θ value | 9.37149e-90 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2C4 | Gene names | Mtmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR1_HUMAN
|
||||||
| NC score | 0.949196 (rank : 11) | θ value | 6.07444e-89 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13613, Q9UBX6, Q9UEM0, Q9UQD5 | Gene names | MTMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR3_MOUSE
|
||||||
| NC score | 0.925299 (rank : 12) | θ value | 2.92676e-75 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.824098 (rank : 13) | θ value | 1.71583e-75 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.698153 (rank : 14) | θ value | 8.06329e-33 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
MTMRD_HUMAN
|
||||||
| NC score | 0.692878 (rank : 15) | θ value | 7.54701e-31 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
LYRIC_HUMAN
|
||||||
| NC score | 0.063181 (rank : 16) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
MYCPP_HUMAN
|
||||||
| NC score | 0.061422 (rank : 17) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.052371 (rank : 18) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.052357 (rank : 19) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
LYRIC_MOUSE
|
||||||
| NC score | 0.051575 (rank : 20) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.051537 (rank : 21) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
DEN4C_HUMAN
|
||||||
| NC score | 0.050774 (rank : 22) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.050223 (rank : 23) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SG16_MOUSE
|
||||||
| NC score | 0.046246 (rank : 24) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.036770 (rank : 25) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.030276 (rank : 26) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.028791 (rank : 27) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
YETS2_MOUSE
|
||||||
| NC score | 0.028512 (rank : 28) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
JHD2A_HUMAN
|
||||||
| NC score | 0.027368 (rank : 29) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
LIGA_MOUSE
|
||||||
| NC score | 0.025648 (rank : 30) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61211, Q3TDE0, Q3THV5, Q3TTP4, Q8C491, Q8CBF1, Q8CC17, Q8R1I9, Q8R3M5 | Gene names | Lgtn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ligatin. | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.024578 (rank : 31) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
HPS5_HUMAN
|
||||||
| NC score | 0.024078 (rank : 32) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPZ3, O95942, Q8N4U0 | Gene names | HPS5, AIBP63, KIAA1017 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein (Alpha-integrin-binding protein 63) (Ruby-eye protein 2 homolog) (Ru2). | |||||
|
HNRPC_HUMAN
|
||||||
| NC score | 0.021801 (rank : 33) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.021409 (rank : 34) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DYH9_HUMAN
|
||||||
| NC score | 0.020119 (rank : 35) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.019966 (rank : 36) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.018723 (rank : 37) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
DMRTA_MOUSE
|
||||||
| NC score | 0.017691 (rank : 38) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFG4 | Gene names | Dmrta1, Dmrt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor A1 (Doublesex- and mab-3-related transcription factor 4). | |||||
|
ELOA3_HUMAN
|
||||||
| NC score | 0.016917 (rank : 39) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
DUS5_HUMAN
|
||||||
| NC score | 0.016574 (rank : 40) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
IPP4_HUMAN
|
||||||
| NC score | 0.016542 (rank : 41) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
PKP4_HUMAN
|
||||||
| NC score | 0.016259 (rank : 42) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
PFD4_HUMAN
|
||||||
| NC score | 0.015874 (rank : 43) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQP4, Q5TD11, Q92779 | Gene names | PFDN4, PFD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prefoldin subunit 4 (Protein C-1). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.015199 (rank : 44) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.014342 (rank : 45) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TACC1_HUMAN
|
||||||
| NC score | 0.014063 (rank : 46) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.013692 (rank : 47) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.013616 (rank : 48) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
HOME1_HUMAN
|
||||||
| NC score | 0.013393 (rank : 49) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1. | |||||
|
DUS2_MOUSE
|
||||||
| NC score | 0.012781 (rank : 50) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
SPC24_MOUSE
|
||||||
| NC score | 0.012755 (rank : 51) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D083, Q3TIF5, Q8CD13 | Gene names | Spbc24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24. | |||||
|
HOME1_MOUSE
|
||||||
| NC score | 0.012740 (rank : 52) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
K1024_HUMAN
|
||||||
| NC score | 0.012466 (rank : 53) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPX6 | Gene names | KIAA1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024. | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.012309 (rank : 54) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
AMBN_HUMAN
|
||||||
| NC score | 0.012036 (rank : 55) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP70, Q9H2X1, Q9H4L1 | Gene names | AMBN | |||
|
Domain Architecture |

|
|||||
| Description | Ameloblastin precursor. | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.011138 (rank : 56) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.011135 (rank : 57) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.010547 (rank : 58) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.010362 (rank : 59) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.010271 (rank : 60) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.008884 (rank : 61) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NEMO_MOUSE
|
||||||
| NC score | 0.008707 (rank : 62) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.008704 (rank : 63) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.008679 (rank : 64) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.007225 (rank : 65) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.007140 (rank : 66) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
PCTK2_MOUSE
|
||||||
| NC score | 0.006718 (rank : 67) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0D0 | Gene names | Pctk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PCTAIRE-2 (EC 2.7.11.22) (PCTAIRE- motif protein kinase 2). | |||||
|
PCTK2_HUMAN
|
||||||
| NC score | 0.006181 (rank : 68) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00537, Q8NEB8 | Gene names | PCTK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PCTAIRE-2 (EC 2.7.11.22) (PCTAIRE- motif protein kinase 2). | |||||
|
UTY_HUMAN
|
||||||
| NC score | 0.005440 (rank : 69) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.004553 (rank : 70) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.004084 (rank : 71) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.004065 (rank : 72) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.003479 (rank : 73) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.003251 (rank : 74) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.003160 (rank : 75) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||