Please be patient as the page loads
|
MTMR3_MOUSE
|
||||||
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |
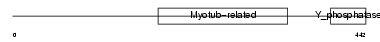
|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MTMR3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.881126 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
MTMR3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
MTMR2_HUMAN
|
||||||
| θ value | 1.49272e-95 (rank : 3) | NC score | 0.940321 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13614, Q9UPS9 | Gene names | MTMR2, KIAA1073 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTMR2_MOUSE
|
||||||
| θ value | 1.65039e-94 (rank : 4) | NC score | 0.940268 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D1, Q8VHA7 | Gene names | Mtmr2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTM1_HUMAN
|
||||||
| θ value | 4.96919e-91 (rank : 5) | NC score | 0.939424 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13496, Q8NEL1 | Gene names | MTM1, CG2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTM1_MOUSE
|
||||||
| θ value | 3.22093e-90 (rank : 6) | NC score | 0.938882 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2C5 | Gene names | Mtm1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTMR1_HUMAN
|
||||||
| θ value | 2.82168e-86 (rank : 7) | NC score | 0.935756 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13613, Q9UBX6, Q9UEM0, Q9UQD5 | Gene names | MTMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR1_MOUSE
|
||||||
| θ value | 3.11973e-85 (rank : 8) | NC score | 0.935489 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2C4 | Gene names | Mtmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR7_HUMAN
|
||||||
| θ value | 2.92676e-75 (rank : 9) | NC score | 0.925299 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR7_MOUSE
|
||||||
| θ value | 2.47765e-74 (rank : 10) | NC score | 0.924092 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR6_HUMAN
|
||||||
| θ value | 5.71191e-71 (rank : 11) | NC score | 0.931983 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y217, Q86TB7, Q86YH4, Q96P80 | Gene names | MTMR6 | |||
|
Domain Architecture |
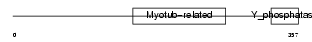
|
|||||
| Description | Myotubularin-related protein 6 (EC 3.1.3.-). | |||||
|
MTMR9_MOUSE
|
||||||
| θ value | 1.2325e-65 (rank : 12) | NC score | 0.928139 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D0, Q80XL4 | Gene names | Mtmr9, Mtmr3 | |||
|
Domain Architecture |
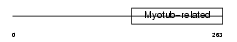
|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR9_HUMAN
|
||||||
| θ value | 2.10232e-65 (rank : 13) | NC score | 0.927915 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QG7, Q52LU3, Q8WW11, Q96QG6, Q9NX50 | Gene names | MTMR9, C8orf9, MTMR8 | |||
|
Domain Architecture |
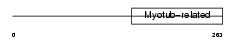
|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 6.15952e-41 (rank : 14) | NC score | 0.713203 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
MTMRD_HUMAN
|
||||||
| θ value | 1.3722e-40 (rank : 15) | NC score | 0.709761 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
PDXL2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 16) | NC score | 0.058080 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
CM007_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.061283 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.011475 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
E41L3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.020542 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.050498 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
CM007_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.054951 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5W0B1, Q8TBY2, Q9H0T2, Q9H8M0 | Gene names | C13orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7. | |||||
|
RAF1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.006892 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99N57, Q91WH1 | Gene names | Raf1, Craf | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
PRDM1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.002380 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.019076 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.012292 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DZIP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.024831 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.016961 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
NUAK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.000732 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.031417 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.012496 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.006457 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | -0.000431 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.018352 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.007730 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
PSD4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.012421 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLR5, Q3TE33, Q3TQF5, Q3UD59, Q3UFH9, Q80V44 | Gene names | Psd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4). | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.019690 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.002850 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.011876 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.000724 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.029314 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
FA53C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.020718 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYF3 | Gene names | FAM53C, C5orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM53C. | |||||
|
SNX15_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.013636 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WE1 | Gene names | Snx15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-15. | |||||
|
TEX2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.014068 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.015780 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.018139 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.012901 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ZBT38_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | -0.000321 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.001354 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.009641 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CHKA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.009113 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35790, Q8NE29 | Gene names | CHKA, CHK, CKI | |||
|
Domain Architecture |

|
|||||
| Description | Choline kinase alpha (EC 2.7.1.32) (CK) (CHETK-alpha). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.003892 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.000121 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PALM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.022025 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75781, O43359, O95673, Q92559, Q9UPJ4, Q9UQS2, Q9UQS3 | Gene names | PALM, KIAA0270 | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.003814 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CLK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | -0.000052 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49759 | Gene names | CLK1, CLK | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK1 (EC 2.7.12.1) (CDC-like kinase 1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.008936 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
FA53C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.017307 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXQ8, Q3U4J2 | Gene names | Fam53c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM53C. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.000759 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
INSM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.003682 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 683 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01101 | Gene names | INSM1, IA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
IRAK4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.000553 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4K2, Q80WW1 | Gene names | Irak4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.004889 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.009409 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.003496 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.005706 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.008425 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.006833 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.008193 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
BNC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.005487 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01954, Q15840 | Gene names | BNC1, BNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
CCDC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.013494 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
ES8L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.004333 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE68, Q71RE2, Q8NC10, Q96BB7, Q9BSQ2, Q9GZQ2, Q9NXH0 | Gene names | EPS8L1, DRC3, EPS8R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
IGHA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.003428 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01878 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig alpha chain C region. | |||||
|
MICA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.002916 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.003692 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PP14C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.007751 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4S0 | Gene names | Ppp1r14c, Kepi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14C (PKC-potentiated PP1 inhibitory protein) (Kinase-enhanced PP1 inhibitor). | |||||
|
PTPRJ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.006691 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTPRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.005583 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.009833 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.002748 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
DEN4C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.057846 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
MYCPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.069441 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.057402 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.058334 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
ZFY21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.059372 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFY21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.060909 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFY28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.053712 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
MTMR3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
MTMR2_HUMAN
|
||||||
| NC score | 0.940321 (rank : 2) | θ value | 1.49272e-95 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13614, Q9UPS9 | Gene names | MTMR2, KIAA1073 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTMR2_MOUSE
|
||||||
| NC score | 0.940268 (rank : 3) | θ value | 1.65039e-94 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D1, Q8VHA7 | Gene names | Mtmr2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTM1_HUMAN
|
||||||
| NC score | 0.939424 (rank : 4) | θ value | 4.96919e-91 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13496, Q8NEL1 | Gene names | MTM1, CG2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTM1_MOUSE
|
||||||
| NC score | 0.938882 (rank : 5) | θ value | 3.22093e-90 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2C5 | Gene names | Mtm1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTMR1_HUMAN
|
||||||
| NC score | 0.935756 (rank : 6) | θ value | 2.82168e-86 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13613, Q9UBX6, Q9UEM0, Q9UQD5 | Gene names | MTMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR1_MOUSE
|
||||||
| NC score | 0.935489 (rank : 7) | θ value | 3.11973e-85 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2C4 | Gene names | Mtmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR6_HUMAN
|
||||||
| NC score | 0.931983 (rank : 8) | θ value | 5.71191e-71 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y217, Q86TB7, Q86YH4, Q96P80 | Gene names | MTMR6 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 6 (EC 3.1.3.-). | |||||
|
MTMR9_MOUSE
|
||||||
| NC score | 0.928139 (rank : 9) | θ value | 1.2325e-65 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D0, Q80XL4 | Gene names | Mtmr9, Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR9_HUMAN
|
||||||
| NC score | 0.927915 (rank : 10) | θ value | 2.10232e-65 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QG7, Q52LU3, Q8WW11, Q96QG6, Q9NX50 | Gene names | MTMR9, C8orf9, MTMR8 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR7_HUMAN
|
||||||
| NC score | 0.925299 (rank : 11) | θ value | 2.92676e-75 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR7_MOUSE
|
||||||
| NC score | 0.924092 (rank : 12) | θ value | 2.47765e-74 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.881126 (rank : 13) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.713203 (rank : 14) | θ value | 6.15952e-41 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
MTMRD_HUMAN
|
||||||
| NC score | 0.709761 (rank : 15) | θ value | 1.3722e-40 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
MYCPP_HUMAN
|
||||||
| NC score | 0.069441 (rank : 16) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.061283 (rank : 17) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
ZFY21_MOUSE
|
||||||
| NC score | 0.060909 (rank : 18) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
ZFY21_HUMAN
|
||||||
| NC score | 0.059372 (rank : 19) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.058334 (rank : 20) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
PDXL2_HUMAN
|
||||||
| NC score | 0.058080 (rank : 21) | θ value | 0.00298849 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
DEN4C_HUMAN
|
||||||
| NC score | 0.057846 (rank : 22) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VZ89, Q6AI48, Q6ZUB3, Q8NCY7, Q9H6N4, Q9NUT1, Q9NWA5 | Gene names | DENND4C, C9orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 4C. | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.057402 (rank : 23) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
CM007_HUMAN
|
||||||
| NC score | 0.054951 (rank : 24) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5W0B1, Q8TBY2, Q9H0T2, Q9H8M0 | Gene names | C13orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7. | |||||
|
ZFY28_HUMAN
|
||||||
| NC score | 0.053712 (rank : 25) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.050498 (rank : 26) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.031417 (rank : 27) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.029314 (rank : 28) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
DZIP1_MOUSE
|
||||||
| NC score | 0.024831 (rank : 29) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
PALM_HUMAN
|
||||||
| NC score | 0.022025 (rank : 30) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75781, O43359, O95673, Q92559, Q9UPJ4, Q9UQS2, Q9UQS3 | Gene names | PALM, KIAA0270 | |||
|
Domain Architecture |

|
|||||
| Description | Paralemmin. | |||||
|
FA53C_HUMAN
|
||||||
| NC score | 0.020718 (rank : 31) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYF3 | Gene names | FAM53C, C5orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM53C. | |||||
|
E41L3_HUMAN
|
||||||
| NC score | 0.020542 (rank : 32) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.019690 (rank : 33) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.019076 (rank : 34) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.018352 (rank : 35) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.018139 (rank : 36) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
FA53C_MOUSE
|
||||||
| NC score | 0.017307 (rank : 37) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BXQ8, Q3U4J2 | Gene names | Fam53c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM53C. | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.016961 (rank : 38) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.015780 (rank : 39) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
TEX2_MOUSE
|
||||||
| NC score | 0.014068 (rank : 40) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
SNX15_MOUSE
|
||||||
| NC score | 0.013636 (rank : 41) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WE1 | Gene names | Snx15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-15. | |||||
|
CCDC4_HUMAN
|
||||||
| NC score | 0.013494 (rank : 42) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.012901 (rank : 43) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.012496 (rank : 44) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
PSD4_MOUSE
|
||||||
| NC score | 0.012421 (rank : 45) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLR5, Q3TE33, Q3TQF5, Q3UD59, Q3UFH9, Q80V44 | Gene names | Psd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.012292 (rank : 46) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.011876 (rank : 47) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.011475 (rank : 48) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.009833 (rank : 49) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.009641 (rank : 50) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.009409 (rank : 51) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
CHKA_HUMAN
|
||||||
| NC score | 0.009113 (rank : 52) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35790, Q8NE29 | Gene names | CHKA, CHK, CKI | |||
|
Domain Architecture |

|
|||||
| Description | Choline kinase alpha (EC 2.7.1.32) (CK) (CHETK-alpha). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.008936 (rank : 53) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.008425 (rank : 54) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.008193 (rank : 55) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
PP14C_MOUSE
|
||||||
| NC score | 0.007751 (rank : 56) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4S0 | Gene names | Ppp1r14c, Kepi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14C (PKC-potentiated PP1 inhibitory protein) (Kinase-enhanced PP1 inhibitor). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.007730 (rank : 57) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
RAF1_MOUSE
|
||||||
| NC score | 0.006892 (rank : 58) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99N57, Q91WH1 | Gene names | Raf1, Craf | |||
|
Domain Architecture |

|
|||||
| Description | RAF proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1) (Raf- 1) (C-RAF) (cRaf). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.006833 (rank : 59) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
PTPRJ_MOUSE
|
||||||
| NC score | 0.006691 (rank : 60) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.006457 (rank : 61) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.005706 (rank : 62) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PTPRO_HUMAN
|
||||||
| NC score | 0.005583 (rank : 63) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
BNC1_HUMAN
|
||||||
| NC score | 0.005487 (rank : 64) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01954, Q15840 | Gene names | BNC1, BNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.004889 (rank : 65) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
ES8L1_HUMAN
|
||||||
| NC score | 0.004333 (rank : 66) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE68, Q71RE2, Q8NC10, Q96BB7, Q9BSQ2, Q9GZQ2, Q9NXH0 | Gene names | EPS8L1, DRC3, EPS8R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.003892 (rank : 67) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.003814 (rank : 68) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.003692 (rank : 69) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
INSM1_HUMAN
|
||||||
| NC score | 0.003682 (rank : 70) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 683 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01101 | Gene names | INSM1, IA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.003496 (rank : 71) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
IGHA_MOUSE
|
||||||
| NC score | 0.003428 (rank : 72) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01878 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig alpha chain C region. | |||||
|
MICA2_HUMAN
|
||||||
| NC score | 0.002916 (rank : 73) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94851, Q7Z3A8 | Gene names | MICAL2, KIAA0750 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.002850 (rank : 74) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.002748 (rank : 75) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
PRDM1_MOUSE
|
||||||
| NC score | 0.002380 (rank : 76) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.001354 (rank : 77) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.000759 (rank : 78) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
NUAK1_HUMAN
|
||||||
| NC score | 0.000732 (rank : 79) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.000724 (rank : 80) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
IRAK4_MOUSE
|
||||||
| NC score | 0.000553 (rank : 81) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4K2, Q80WW1 | Gene names | Irak4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-1 receptor-associated kinase 4 (EC 2.7.11.1) (IRAK-4). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.000121 (rank : 82) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
CLK1_HUMAN
|
||||||
| NC score | -0.000052 (rank : 83) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49759 | Gene names | CLK1, CLK | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase CLK1 (EC 2.7.12.1) (CDC-like kinase 1). | |||||
|
ZBT38_HUMAN
|
||||||
| NC score | -0.000321 (rank : 84) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NAP3 | Gene names | ZBTB38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 38. | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.000431 (rank : 85) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||