Please be patient as the page loads
|
E41L3_HUMAN
|
||||||
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
E41L3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.980983 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
E41L1_HUMAN
|
||||||
| θ value | 1.72287e-176 (rank : 3) | NC score | 0.980953 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_MOUSE
|
||||||
| θ value | 5.01279e-176 (rank : 4) | NC score | 0.986219 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 3.72618e-163 (rank : 5) | NC score | 0.973318 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 9.49765e-159 (rank : 6) | NC score | 0.952375 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
41_HUMAN
|
||||||
| θ value | 1.20145e-153 (rank : 7) | NC score | 0.981257 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
41_MOUSE
|
||||||
| θ value | 4.88915e-147 (rank : 8) | NC score | 0.979422 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 7.68416e-84 (rank : 9) | NC score | 0.678999 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
E41L5_HUMAN
|
||||||
| θ value | 8.51556e-75 (rank : 10) | NC score | 0.965636 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
E41LB_MOUSE
|
||||||
| θ value | 1.35953e-72 (rank : 11) | NC score | 0.965511 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
E41LB_HUMAN
|
||||||
| θ value | 6.7473e-72 (rank : 12) | NC score | 0.959377 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 1.27248e-70 (rank : 13) | NC score | 0.802663 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
E41LA_HUMAN
|
||||||
| θ value | 3.70236e-70 (rank : 14) | NC score | 0.958406 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 2.3998e-69 (rank : 15) | NC score | 0.770864 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 1.31681e-67 (rank : 16) | NC score | 0.778548 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
E41LA_MOUSE
|
||||||
| θ value | 1.11475e-66 (rank : 17) | NC score | 0.958094 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |
|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
FRMD5_MOUSE
|
||||||
| θ value | 1.11475e-66 (rank : 18) | NC score | 0.963951 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD5_HUMAN
|
||||||
| θ value | 1.2325e-65 (rank : 19) | NC score | 0.961235 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
PTN3_HUMAN
|
||||||
| θ value | 4.24587e-58 (rank : 20) | NC score | 0.649843 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | 1.79215e-40 (rank : 21) | NC score | 0.621864 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 22) | NC score | 0.620046 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 23) | NC score | 0.737000 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 4.27553e-34 (rank : 24) | NC score | 0.741600 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 25) | NC score | 0.680190 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 26) | NC score | 0.672830 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 1.16434e-31 (rank : 27) | NC score | 0.722230 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 28) | NC score | 0.718479 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
MERL_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 29) | NC score | 0.776156 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
MERL_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 30) | NC score | 0.776577 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
PTN14_MOUSE
|
||||||
| θ value | 4.14116e-29 (rank : 31) | NC score | 0.600930 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | 1.2049e-28 (rank : 32) | NC score | 0.593210 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
MYLIP_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 33) | NC score | 0.846151 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 34) | NC score | 0.861451 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 35) | NC score | 0.481767 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 36) | NC score | 0.472655 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
FRM4B_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 37) | NC score | 0.789774 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
FRM4B_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 38) | NC score | 0.787501 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
FRM4A_MOUSE
|
||||||
| θ value | 1.02238e-19 (rank : 39) | NC score | 0.778650 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FRM4A_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 40) | NC score | 0.777230 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
TLN2_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 41) | NC score | 0.408703 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
TLN2_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 42) | NC score | 0.412279 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
FRMD6_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 43) | NC score | 0.659333 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |
|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
FRMD6_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 44) | NC score | 0.650338 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |
|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
FRMD1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 45) | NC score | 0.638788 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 46) | NC score | 0.417628 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 47) | NC score | 0.414456 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
MTMR3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 48) | NC score | 0.020542 (rank : 142) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |
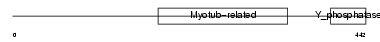
|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
KIF3A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 49) | NC score | 0.024503 (rank : 140) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 50) | NC score | 0.027052 (rank : 137) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
ALS2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.039458 (rank : 127) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.009855 (rank : 163) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
KIRR1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.011143 (rank : 159) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W68, Q8CIJ4, Q8CJ59, Q923L4 | Gene names | Kirrel1, Kirrel, Neph1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.053543 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.031747 (rank : 133) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
POK17_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.018555 (rank : 147) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63136 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pol protein [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.046767 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
POK5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.018308 (rank : 150) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63132 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pol protein (HERV-K113 Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
POK6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.018316 (rank : 149) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63133 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pol protein (HERV-K115 Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
ACHA4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.003953 (rank : 177) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43681, Q4JGR7 | Gene names | CHRNA4, NACRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-4 precursor. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.016064 (rank : 154) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.012176 (rank : 156) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
EP15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.019073 (rank : 145) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.011484 (rank : 158) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
M3K12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | -0.001921 (rank : 183) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.033851 (rank : 131) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
CJ068_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.035652 (rank : 130) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
KIRR1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.009363 (rank : 165) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J84, Q5XKC6, Q7Z696, Q7Z7N8, Q8TB15, Q9H9N1, Q9NVA5 | Gene names | KIRREL, NEPH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
REST_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.027649 (rank : 136) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.012532 (rank : 155) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | -0.001934 (rank : 184) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.008503 (rank : 169) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.024423 (rank : 141) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.010909 (rank : 160) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.037725 (rank : 129) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
PKHC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.040466 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CIB5, Q8C542, Q8K035 | Gene names | Plekhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1. | |||||
|
TSH3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.006307 (rank : 172) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.019812 (rank : 143) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
IKKB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | -0.002961 (rank : 185) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14920, O75327 | Gene names | IKBKB, IKKB | |||
|
Domain Architecture |
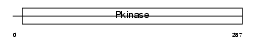
|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.009974 (rank : 162) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PKHC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.042273 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96AC1, Q14840, Q86TY7 | Gene names | PLEKHC1, KIND2, MIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1 (Kindlin-2) (Mitogen-inducible gene 2 protein) (Mig-2). | |||||
|
REST_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.032445 (rank : 132) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.016787 (rank : 152) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.007931 (rank : 170) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.018580 (rank : 146) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.029452 (rank : 134) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.018419 (rank : 148) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
HAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.039089 (rank : 128) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
MORC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.025003 (rank : 139) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14149, Q9UEZ2 | Gene names | MORC3, KIAA0136, ZCWCC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 3 (Zinc finger CW-type coiled- coil domain protein 3). | |||||
|
POK3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.016232 (rank : 153) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WJR5, O15312 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pol protein (HERV-K(C19) Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
ZHX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.009764 (rank : 164) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
ANR38_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.002425 (rank : 180) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T7N3, Q6P9A0, Q86T71, Q86VE6, Q8NAX3 | Gene names | ANKRD38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
LAMC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.006876 (rank : 171) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
M3K12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | -0.003548 (rank : 187) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
MKS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.008666 (rank : 167) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SW45, Q3V3W3 | Gene names | Mks1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Meckel syndrome type 1 protein homolog. | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.011861 (rank : 157) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.017721 (rank : 151) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
ZHX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.009086 (rank : 166) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
ZN560_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | -0.005281 (rank : 188) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96MR9, Q495S9, Q495T1 | Gene names | ZNF560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 560. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.008604 (rank : 168) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.019643 (rank : 144) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RAE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.004457 (rank : 174) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZD5, Q8CH16 | Gene names | Chml, Rep2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.004211 (rank : 176) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | -0.001600 (rank : 182) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANR35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.000730 (rank : 181) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.026130 (rank : 138) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
ERCC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.010140 (rank : 161) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.002855 (rank : 179) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.029080 (rank : 135) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.004287 (rank : 175) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
NRK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | -0.003152 (rank : 186) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
TSH3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.003917 (rank : 178) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63HK5, Q9H0G6, Q9P254 | Gene names | TSHZ3, KIAA1474, TSH3, ZNF537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.006257 (rank : 173) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ARHG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.050777 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
ARHG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.052013 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
CA106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.084749 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
CA106_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.083605 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
CD45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.098919 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
CD45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.103952 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.059049 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.057136 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.050977 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
FGD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.055516 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
FGD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.053473 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
FGD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.051941 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
FGD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.060927 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.061586 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
FGD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.064385 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
FGD5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.060673 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
FGD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.063473 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.062436 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.056460 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.052634 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.059509 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
KRIT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.083003 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |
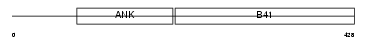
|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
KRIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.077044 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6S5J6, Q6VSV2, Q7TPR8, Q8C9Q6, Q9EPY2, Q9ERH0 | Gene names | Krit1, Ccm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein homolog). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.056216 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PTN11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.102459 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.102450 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
PTN12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.109094 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.108787 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
PTN18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.117921 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |
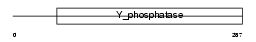
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PTN18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.114401 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.117876 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.118248 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
PTN22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.110729 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.109030 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.102992 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.101358 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.117882 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.119137 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
PTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.102473 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.102478 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.108923 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.108896 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PTN7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.109164 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
PTN7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.110328 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
PTN9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.113953 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.113586 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTPR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.106930 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
PTPR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.106756 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTPRA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.095175 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.095139 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
PTPRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.100534 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.067627 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.065206 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.095515 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.095033 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.065977 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.085389 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.085559 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRJ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.102895 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTPRJ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.103133 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTPRK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.086070 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.086776 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.081102 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.081414 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.106493 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.106096 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTPRO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.107185 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPRR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.104368 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
PTPRR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.104711 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
PTPRS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.068709 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.083240 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.083196 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
PTPRU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.080940 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.102046 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.085244 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
E41L3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9Y2J2, O95713, Q9BRP5 | Gene names | EPB41L3, DAL1, KIAA0987 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1). | |||||
|
E41L1_MOUSE
|
||||||
| NC score | 0.986219 (rank : 2) | θ value | 5.01279e-176 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.981257 (rank : 3) | θ value | 1.20145e-153 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.980983 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.980953 (rank : 5) | θ value | 1.72287e-176 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.979422 (rank : 6) | θ value | 4.88915e-147 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.973318 (rank : 7) | θ value | 3.72618e-163 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
E41L5_HUMAN
|
||||||
| NC score | 0.965636 (rank : 8) | θ value | 8.51556e-75 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCM4, Q9H975 | Gene names | EPB41L5, KIAA1548 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 5. | |||||
|
E41LB_MOUSE
|
||||||
| NC score | 0.965511 (rank : 9) | θ value | 1.35953e-72 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JMC8 | Gene names | Epb41l4b, Ehm2, Epb4.1l4b | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2). | |||||
|
FRMD5_MOUSE
|
||||||
| NC score | 0.963951 (rank : 10) | θ value | 1.11475e-66 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P5H6, Q6PFH6, Q8C0K0 | Gene names | Frmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
FRMD5_HUMAN
|
||||||
| NC score | 0.961235 (rank : 11) | θ value | 1.2325e-65 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6J6, Q8NBG4 | Gene names | FRMD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 5. | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.959377 (rank : 12) | θ value | 6.7473e-72 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
E41LA_HUMAN
|
||||||
| NC score | 0.958406 (rank : 13) | θ value | 3.70236e-70 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCS5 | Gene names | EPB41L4A, EPB41L4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41LA_MOUSE
|
||||||
| NC score | 0.958094 (rank : 14) | θ value | 1.11475e-66 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P52963 | Gene names | Epb41l4a, Epb4.1l4, Epb4.1l4a, Epb41l4 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4A (Protein NBL4). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.952375 (rank : 15) | θ value | 9.49765e-159 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MYLIP_MOUSE
|
||||||
| NC score | 0.861451 (rank : 16) | θ value | 3.87602e-27 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_HUMAN
|
||||||
| NC score | 0.846151 (rank : 17) | θ value | 1.73987e-27 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.802663 (rank : 18) | θ value | 1.27248e-70 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
FRM4B_HUMAN
|
||||||
| NC score | 0.789774 (rank : 19) | θ value | 3.75424e-22 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
FRM4B_MOUSE
|
||||||
| NC score | 0.787501 (rank : 20) | θ value | 4.15078e-21 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
FRM4A_MOUSE
|
||||||
| NC score | 0.778650 (rank : 21) | θ value | 1.02238e-19 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BIE6, Q6PDJ4, Q8CHA6 | Gene names | Frmd4a, Frmd4, Kiaa1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.778548 (rank : 22) | θ value | 1.31681e-67 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FRM4A_HUMAN
|
||||||
| NC score | 0.777230 (rank : 23) | θ value | 1.74391e-19 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9P2Q2 | Gene names | FRMD4A, FRMD4, KIAA1294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4A. | |||||
|
MERL_MOUSE
|
||||||
| NC score | 0.776577 (rank : 24) | θ value | 2.59387e-31 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P46662, Q8BR03 | Gene names | Nf2, Nf-2 | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin). | |||||
|
MERL_HUMAN
|
||||||
| NC score | 0.776156 (rank : 25) | θ value | 1.98606e-31 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P35240, O95683, Q8WUJ2, Q969N0, Q969Q3, Q96T30, Q96T31, Q96T32, Q96T33, Q9BTW3, Q9UNG9, Q9UNH3, Q9UNH4 | Gene names | NF2, SCH | |||
|
Domain Architecture |

|
|||||
| Description | Merlin (Moesin-ezrin-radixin-like protein) (Neurofibromin-2) (Schwannomin) (Schwannomerlin). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.770864 (rank : 26) | θ value | 2.3998e-69 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.741600 (rank : 27) | θ value | 4.27553e-34 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.737000 (rank : 28) | θ value | 1.46948e-34 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.722230 (rank : 29) | θ value | 1.16434e-31 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.718479 (rank : 30) | θ value | 1.16434e-31 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.680190 (rank : 31) | θ value | 3.06405e-32 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.678999 (rank : 32) | θ value | 7.68416e-84 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.672830 (rank : 33) | θ value | 5.22648e-32 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
FRMD6_MOUSE
|
||||||
| NC score | 0.659333 (rank : 34) | θ value | 2.61198e-07 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C0V9, Q8BW34 | Gene names | Frmd6 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6. | |||||
|
FRMD6_HUMAN
|
||||||
| NC score | 0.650338 (rank : 35) | θ value | 3.41135e-07 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96NE9, Q5HYF2, Q8N2X1, Q8WUH7 | Gene names | FRMD6, C14orf31 | |||
|
Domain Architecture |

|
|||||
| Description | FERM domain-containing protein 6 (Willin). | |||||
|
PTN3_HUMAN
|
||||||
| NC score | 0.649843 (rank : 36) | θ value | 4.24587e-58 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
FRMD1_HUMAN
|
||||||
| NC score | 0.638788 (rank : 37) | θ value | 4.45536e-07 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N878, Q5SZU7, Q9UFB0 | Gene names | FRMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 1. | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.621864 (rank : 38) | θ value | 1.79215e-40 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.620046 (rank : 39) | θ value | 2.34062e-40 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
PTN14_MOUSE
|
||||||
| NC score | 0.600930 (rank : 40) | θ value | 4.14116e-29 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62130 | Gene names | Ptpn14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP36). | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.593210 (rank : 41) | θ value | 1.2049e-28 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.481767 (rank : 42) | θ value | 1.24688e-25 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.472655 (rank : 43) | θ value | 2.12685e-25 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.417628 (rank : 44) | θ value | 4.45536e-07 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.414456 (rank : 45) | θ value | 4.45536e-07 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN2_MOUSE
|
||||||
| NC score | 0.412279 (rank : 46) | θ value | 1.99992e-07 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
TLN2_HUMAN
|
||||||
| NC score | 0.408703 (rank : 47) | θ value | 1.17247e-07 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
PTN2_MOUSE
|
||||||
| NC score | 0.119137 (rank : 48) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
PTN1_MOUSE
|
||||||
| NC score | 0.118248 (rank : 49) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
PTN18_HUMAN
|
||||||
| NC score | 0.117921 (rank : 50) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99952 | Gene names | PTPN18, BDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Brain-derived phosphatase). | |||||
|
PTN2_HUMAN
|
||||||
| NC score | 0.117882 (rank : 51) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17706, Q96HR2 | Gene names | PTPN2, PTPT | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (T-cell protein-tyrosine phosphatase) (TCPTP). | |||||
|
PTN1_HUMAN
|
||||||
| NC score | 0.117876 (rank : 52) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18031, Q5TGD8, Q9BQV9, Q9NQQ4 | Gene names | PTPN1, PTP1B | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B). | |||||
|
PTN18_MOUSE
|
||||||
| NC score | 0.114401 (rank : 53) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
PTN9_HUMAN
|
||||||
| NC score | 0.113953 (rank : 54) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_MOUSE
|
||||||
| NC score | 0.113586 (rank : 55) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN22_HUMAN
|
||||||
| NC score | 0.110729 (rank : 56) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2R2, O95063, O95064 | Gene names | PTPN22, PTPN8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP) (Lymphoid phosphatase) (LyP). | |||||
|
PTN7_MOUSE
|
||||||
| NC score | 0.110328 (rank : 57) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUM3 | Gene names | Ptpn7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48). | |||||
|
PTN7_HUMAN
|
||||||
| NC score | 0.109164 (rank : 58) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
PTN12_HUMAN
|
||||||
| NC score | 0.109094 (rank : 59) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.109030 (rank : 60) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
PTN6_HUMAN
|
||||||
| NC score | 0.108923 (rank : 61) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
PTN6_MOUSE
|
||||||
| NC score | 0.108896 (rank : 62) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29351, O35128, Q63872, Q63873, Q63874, Q921G3, Q9QVA6, Q9QVA7, Q9QVA8, Q9R0V6 | Gene names | Ptpn6, Hcp, Hcph, Ptp1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (70Z-SHP) (SH-PTP1) (SHP-1) (PTPTY-42). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.108787 (rank : 63) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
PTPRO_HUMAN
|
||||||
| NC score | 0.107185 (rank : 64) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
PTPR2_HUMAN
|
||||||
| NC score | 0.106930 (rank : 65) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92932, Q8N4I5, Q92662 | Gene names | PTPRN2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (Islet cell autoantigen-related protein) (ICAAR) (IAR) (Phogrin). | |||||
|
PTPR2_MOUSE
|
||||||
| NC score | 0.106756 (rank : 66) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P80560, O09134, P70328 | Gene names | Ptprn2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase N2 precursor (EC 3.1.3.48) (R-PTP-N2) (PTP IA-2beta) (Protein tyrosine phosphatase-NP) (PTP-NP). | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.106493 (rank : 67) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.106096 (rank : 68) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PTPRR_MOUSE
|
||||||
| NC score | 0.104711 (rank : 69) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62132, Q64491, Q64492, Q9QUH9 | Gene names | Ptprr, Ptp13 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine-phosphatase SL) (Phosphotyrosine phosphatase 13). | |||||
|
PTPRR_HUMAN
|
||||||
| NC score | 0.104368 (rank : 70) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.103952 (rank : 71) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
PTPRJ_MOUSE
|
||||||
| NC score | 0.103133 (rank : 72) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.102992 (rank : 73) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PTPRJ_HUMAN
|
||||||
| NC score | 0.102895 (rank : 74) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTN5_MOUSE
|
||||||
| NC score | 0.102478 (rank : 75) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN5_HUMAN
|
||||||
| NC score | 0.102473 (rank : 76) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54829, Q8N2A1 | Gene names | PTPN5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
PTN11_HUMAN
|
||||||
| NC score | 0.102459 (rank : 77) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06124, Q96HD7 | Gene names | PTPN11, PTP2C, SHPTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase 2C) (PTP-2C) (PTP-1D) (SH-PTP3) (SH- PTP2) (SHP-2) (Shp2). | |||||
|
PTN11_MOUSE
|
||||||
| NC score | 0.102450 (rank : 78) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35235, Q3TQ84, Q64509, Q6PCL5 | Gene names | Ptpn11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 11 (EC 3.1.3.48) (Protein-tyrosine phosphatase SYP) (SH-PTP2) (SHP-2) (Shp2). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.102046 (rank : 79) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.101358 (rank : 80) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTPRB_HUMAN
|
||||||
| NC score | 0.100534 (rank : 81) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P23467 | Gene names | PTPRB, PTPB | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase beta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase beta) (R-PTP-beta). | |||||
|
CD45_HUMAN
|
||||||
| NC score | 0.098919 (rank : 82) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08575, Q16614, Q9H0Y6 | Gene names | PTPRC, CD45 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (CD45 antigen) (T200). | |||||
|
PTPRE_HUMAN
|
||||||
| NC score | 0.095515 (rank : 83) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRA_HUMAN
|
||||||
| NC score | 0.095175 (rank : 84) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18433, Q14513, Q7Z2I2, Q96TD9 | Gene names | PTPRA, PTPA | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha). | |||||
|
PTPRA_MOUSE
|
||||||
| NC score | 0.095139 (rank : 85) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
PTPRE_MOUSE
|
||||||
| NC score | 0.095033 (rank : 86) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
PTPRK_MOUSE
|
||||||
| NC score | 0.086776 (rank : 87) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35822 | Gene names | Ptprk, Ptpk | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRK_HUMAN
|
||||||
| NC score | 0.086070 (rank : 88) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15262, Q14763 | Gene names | PTPRK, PTPK | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase kappa precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase kappa) (R-PTP-kappa). | |||||
|
PTPRG_MOUSE
|
||||||
| NC score | 0.085559 (rank : 89) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.085389 (rank : 90) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.085244 (rank : 91) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
CA106_HUMAN
|
||||||
| NC score | 0.084749 (rank : 92) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
CA106_MOUSE
|
||||||
| NC score | 0.083605 (rank : 93) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TN12, Q7TN27, Q8BGK3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106 homolog. | |||||
|
PTPRT_HUMAN
|
||||||
| NC score | 0.083240 (rank : 94) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14522, O43655, O75664, Q5W0X9, Q5W0Y1, Q9BR24, Q9BR28, Q9H0Y8, Q9NTL1, Q9NU72, Q9UBD2, Q9UJL7 | Gene names | PTPRT, KIAA0283 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho). | |||||
|
PTPRT_MOUSE
|
||||||
| NC score | 0.083196 (rank : 95) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99M80, Q99M81, Q99M82, Q9JIZ1, Q9JIZ2, Q9JKC2, Q9JLP0 | Gene names | Ptprt | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase T precursor (EC 3.1.3.48) (R-PTP-T) (RPTP-rho) (mRPTPrho) (RPTPmam4). | |||||
|
KRIT1_HUMAN
|
||||||
| NC score | 0.083003 (rank : 96) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |

|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
PTPRM_MOUSE
|
||||||
| NC score | 0.081414 (rank : 97) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28828 | Gene names | Ptprm | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRM_HUMAN
|
||||||
| NC score | 0.081102 (rank : 98) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P28827 | Gene names | PTPRM, PTPRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase mu precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase mu) (R-PTP-mu). | |||||
|
PTPRU_HUMAN
|
||||||
| NC score | 0.080940 (rank : 99) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92729 | Gene names | PTPRU, PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase U precursor (EC 3.1.3.48) (R-PTP-U) (Protein-tyrosine phosphatase J) (PTP-J) (Pancreatic carcinoma phosphatase 2) (PCP-2). | |||||
|
KRIT1_MOUSE
|
||||||
| NC score | 0.077044 (rank : 100) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6S5J6, Q6VSV2, Q7TPR8, Q8C9Q6, Q9EPY2, Q9ERH0 | Gene names | Krit1, Ccm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein homolog). | |||||
|
PTPRS_HUMAN
|
||||||
| NC score | 0.068709 (rank : 101) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13332, O75255, O75870, Q15718, Q16341 | Gene names | PTPRS | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase S precursor (EC 3.1.3.48) (R-PTP-S) (Protein-tyrosine phosphatase sigma) (R-PTP-sigma). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.067627 (rank : 102) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRF_HUMAN
|
||||||
| NC score | 0.065977 (rank : 103) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10586 | Gene names | PTPRF, LAR | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase F precursor (EC 3.1.3.48) (LAR protein) (Leukocyte antigen related). | |||||
|
PTPRD_MOUSE
|
||||||
| NC score | 0.065206 (rank : 104) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.064385 (rank : 105) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.063473 (rank : 106) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.062436 (rank : 107) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.061586 (rank : 108) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
FGD4_HUMAN
|
||||||
| NC score | 0.060927 (rank : 109) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96M96, Q6ULS2, Q8TCP6 | Gene names | FGD4, FRABP, ZFYVE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein) (Zinc finger FYVE domain-containing protein 6). | |||||
|
FGD5_MOUSE
|
||||||
| NC score | 0.060673 (rank : 110) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.059509 (rank : 111) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.059049 (rank : 112) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.057136 (rank : 113) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.056460 (rank : 114) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.056216 (rank : 115) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
FGD2_MOUSE
|
||||||
| NC score | 0.055516 (rank : 116) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BY35, O88841, Q7TSE3, Q8VDH4 | Gene names | Fgd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.053543 (rank : 117) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.053473 (rank : 118) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.052634 (rank : 119) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
ARHG4_MOUSE
|
||||||
| NC score | 0.052013 (rank : 120) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TNR9, Q80TJ6 | Gene names | Arhgef4, Kiaa1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
FGD3_MOUSE
|
||||||
| NC score | 0.051941 (rank : 121) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
FGD2_HUMAN
|
||||||
| NC score | 0.050977 (rank : 122) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z6J4, Q5T8I1, Q6P6A8, Q6ZNL5, Q8IZ32, Q8N868, Q9H7M2 | Gene names | FGD2, ZFYVE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 2 (Zinc finger FYVE domain-containing protein 4). | |||||
|
ARHG4_HUMAN
|
||||||
| NC score | 0.050777 (rank : 123) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NR80, Q9HDC6, Q9UPP0 | Gene names | ARHGEF4, KIAA1112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 4 (APC-stimulated guanine nucleotide exchange factor) (Asef). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.046767 (rank : 124) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PKHC1_HUMAN
|
||||||
| NC score | 0.042273 (rank : 125) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96AC1, Q14840, Q86TY7 | Gene names | PLEKHC1, KIND2, MIG2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1 (Kindlin-2) (Mitogen-inducible gene 2 protein) (Mig-2). | |||||
|
PKHC1_MOUSE
|
||||||
| NC score | 0.040466 (rank : 126) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CIB5, Q8C542, Q8K035 | Gene names | Plekhc1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family C member 1. | |||||
|
ALS2_HUMAN
|
||||||
| NC score | 0.039458 (rank : 127) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96Q42, Q8N1E0, Q96PC4, Q96Q41, Q9H973, Q9HCK9 | Gene names | ALS2, KIAA1563 | |||
|
Domain Architecture |

|
|||||
| Description | Alsin (Amyotrophic lateral sclerosis protein 2). | |||||
|
HAP1_HUMAN
|
||||||
| NC score | 0.039089 (rank : 128) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54257, O75358, Q9H4G3, Q9HA98, Q9NY90 | Gene names | HAP1, HLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1) (Neuroan 1). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.037725 (rank : 129) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CJ068_HUMAN
|
||||||
| NC score | 0.035652 (rank : 130) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.033851 (rank : 131) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.032445 (rank : 132) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.031747 (rank : 133) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.029452 (rank : 134) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.029080 (rank : 135) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.027649 (rank : 136) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.027052 (rank : 137) | θ value | 0.125558 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.026130 (rank : 138) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MORC3_HUMAN
|
||||||
| NC score | 0.025003 (rank : 139) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14149, Q9UEZ2 | Gene names | MORC3, KIAA0136, ZCWCC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 3 (Zinc finger CW-type coiled- coil domain protein 3). | |||||
|
KIF3A_HUMAN
|
||||||
| NC score | 0.024503 (rank : 140) | θ value | 0.0563607 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.024423 (rank : 141) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
MTMR3_MOUSE
|
||||||
| NC score | 0.020542 (rank : 142) | θ value | 0.0330416 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.019812 (rank : 143) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.019643 (rank : 144) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.019073 (rank : 145) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.018580 (rank : 146) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
POK17_HUMAN
|
||||||
| NC score | 0.018555 (rank : 147) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63136 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pol protein [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.018419 (rank : 148) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
POK6_HUMAN
|
||||||
| NC score | 0.018316 (rank : 149) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63133 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pol protein (HERV-K115 Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
POK5_HUMAN
|
||||||
| NC score | 0.018308 (rank : 150) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63132 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pol protein (HERV-K113 Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.017721 (rank : 151) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.016787 (rank : 152) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
POK3_HUMAN
|
||||||
| NC score | 0.016232 (rank : 153) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WJR5, O15312 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pol protein (HERV-K(C19) Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.016064 (rank : 154) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.012532 (rank : 155) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.012176 (rank : 156) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.011861 (rank : 157) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.011484 (rank : 158) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
KIRR1_MOUSE
|
||||||
| NC score | 0.011143 (rank : 159) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W68, Q8CIJ4, Q8CJ59, Q923L4 | Gene names | Kirrel1, Kirrel, Neph1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.010909 (rank : 160) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ERCC2_HUMAN
|
||||||
| NC score | 0.010140 (rank : 161) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.009974 (rank : 162) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.009855 (rank : 163) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZHX1_MOUSE
|
||||||
| NC score | 0.009764 (rank : 164) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
KIRR1_HUMAN
|
||||||
| NC score | 0.009363 (rank : 165) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J84, Q5XKC6, Q7Z696, Q7Z7N8, Q8TB15, Q9H9N1, Q9NVA5 | Gene names | KIRREL, NEPH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kin of IRRE-like protein 1 precursor (Kin of irregular chiasm-like protein 1) (Nephrin-like protein 1). | |||||
|
ZHX1_HUMAN
|
||||||
| NC score | 0.009086 (rank : 166) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
MKS1_MOUSE
|
||||||
| NC score | 0.008666 (rank : 167) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SW45, Q3V3W3 | Gene names | Mks1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Meckel syndrome type 1 protein homolog. | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.008604 (rank : 168) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.008503 (rank : 169) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.007931 (rank : 170) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
LAMC1_HUMAN
|
||||||
| NC score | 0.006876 (rank : 171) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
TSH3_MOUSE
|
||||||
| NC score | 0.006307 (rank : 172) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.006257 (rank : 173) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
RAE2_MOUSE
|
||||||
| NC score | 0.004457 (rank : 174) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZD5, Q8CH16 | Gene names | Chml, Rep2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.004287 (rank : 175) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.004211 (rank : 176) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
ACHA4_HUMAN
|
||||||
| NC score | 0.003953 (rank : 177) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43681, Q4JGR7 | Gene names | CHRNA4, NACRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-4 precursor. | |||||
|
TSH3_HUMAN
|
||||||
| NC score | 0.003917 (rank : 178) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63HK5, Q9H0G6, Q9P254 | Gene names | TSHZ3, KIAA1474, TSH3, ZNF537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.002855 (rank : 179) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ANR38_HUMAN
|
||||||
| NC score | 0.002425 (rank : 180) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T7N3, Q6P9A0, Q86T71, Q86VE6, Q8NAX3 | Gene names | ANKRD38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.000730 (rank : 181) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
ANK1_MOUSE
|
||||||
| NC score | -0.001600 (rank : 182) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
M3K12_HUMAN
|
||||||
| NC score | -0.001921 (rank : 183) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12852, Q86VQ5, Q8WY25 | Gene names | MAP3K12, ZPK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | -0.001934 (rank : 184) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
IKKB_HUMAN
|
||||||
| NC score | -0.002961 (rank : 185) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14920, O75327 | Gene names | IKBKB, IKKB | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
NRK_HUMAN
|
||||||
| NC score | -0.003152 (rank : 186) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
M3K12_MOUSE
|
||||||
| NC score | -0.003548 (rank : 187) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60700, P70286, Q3TLL7, Q8C4N7, Q8CBX3, Q8CDL6 | Gene names | Map3k12, Zpk | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 12 (EC 2.7.11.25) (Mixed lineage kinase) (Leucine-zipper protein kinase) (ZPK) (Dual leucine zipper bearing kinase) (DLK) (MAPK-upstream kinase) (MUK). | |||||
|
ZN560_HUMAN
|
||||||
| NC score | -0.005281 (rank : 188) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96MR9, Q495S9, Q495T1 | Gene names | ZNF560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 560. | |||||