Please be patient as the page loads
|
MTM1_MOUSE
|
||||||
| SwissProt Accessions | Q9Z2C5 | Gene names | Mtm1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MTM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999642 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13496, Q8NEL1 | Gene names | MTM1, CG2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z2C5 | Gene names | Mtm1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTMR1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.993141 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13613, Q9UBX6, Q9UEM0, Q9UQD5 | Gene names | MTMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR1_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993450 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2C4 | Gene names | Mtmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR2_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.995160 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13614, Q9UPS9 | Gene names | MTMR2, KIAA1073 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTMR2_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.995130 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2D1, Q8VHA7 | Gene names | Mtmr2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTMR6_HUMAN
|
||||||
| θ value | 7.64859e-100 (rank : 7) | NC score | 0.963961 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y217, Q86TB7, Q86YH4, Q96P80 | Gene names | MTMR6 | |||
|
Domain Architecture |
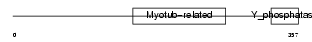
|
|||||
| Description | Myotubularin-related protein 6 (EC 3.1.3.-). | |||||
|
MTMR7_HUMAN
|
||||||
| θ value | 2.38316e-93 (rank : 8) | NC score | 0.951452 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR3_HUMAN
|
||||||
| θ value | 4.96919e-91 (rank : 9) | NC score | 0.834850 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
MTMR7_MOUSE
|
||||||
| θ value | 8.47616e-91 (rank : 10) | NC score | 0.950011 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR3_MOUSE
|
||||||
| θ value | 3.22093e-90 (rank : 11) | NC score | 0.938882 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |
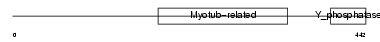
|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
MTMR9_HUMAN
|
||||||
| θ value | 3.56943e-81 (rank : 12) | NC score | 0.956604 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QG7, Q52LU3, Q8WW11, Q96QG6, Q9NX50 | Gene names | MTMR9, C8orf9, MTMR8 | |||
|
Domain Architecture |
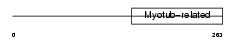
|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR9_MOUSE
|
||||||
| θ value | 4.36332e-79 (rank : 13) | NC score | 0.955880 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D0, Q80XL4 | Gene names | Mtmr9, Mtmr3 | |||
|
Domain Architecture |
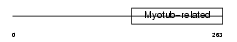
|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMRD_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 14) | NC score | 0.700854 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 5.584e-34 (rank : 15) | NC score | 0.701921 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
DUS4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.046490 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.046551 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.063892 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.050393 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.049212 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.049247 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
PTPRJ_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.011745 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
KV1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.002267 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01593 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region AG. | |||||
|
PTPRJ_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.011063 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.004023 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
DUS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.033928 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.037383 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.008717 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
DUS16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.032528 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
AP3M2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.007078 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
KV1O_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.001159 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01607 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Rei. | |||||
|
SMYD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.011810 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
AP3M2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.006541 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
DUS5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.037605 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.009303 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
SORL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.000561 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
MYCPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.059975 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.050289 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
MTM1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z2C5 | Gene names | Mtm1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTM1_HUMAN
|
||||||
| NC score | 0.999642 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13496, Q8NEL1 | Gene names | MTM1, CG2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin (EC 3.1.3.48). | |||||
|
MTMR2_HUMAN
|
||||||
| NC score | 0.995160 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13614, Q9UPS9 | Gene names | MTMR2, KIAA1073 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTMR2_MOUSE
|
||||||
| NC score | 0.995130 (rank : 4) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2D1, Q8VHA7 | Gene names | Mtmr2 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 2 (EC 3.1.3.-). | |||||
|
MTMR1_MOUSE
|
||||||
| NC score | 0.993450 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2C4 | Gene names | Mtmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR1_HUMAN
|
||||||
| NC score | 0.993141 (rank : 6) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13613, Q9UBX6, Q9UEM0, Q9UQD5 | Gene names | MTMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MTMR6_HUMAN
|
||||||
| NC score | 0.963961 (rank : 7) | θ value | 7.64859e-100 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y217, Q86TB7, Q86YH4, Q96P80 | Gene names | MTMR6 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 6 (EC 3.1.3.-). | |||||
|
MTMR9_HUMAN
|
||||||
| NC score | 0.956604 (rank : 8) | θ value | 3.56943e-81 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QG7, Q52LU3, Q8WW11, Q96QG6, Q9NX50 | Gene names | MTMR9, C8orf9, MTMR8 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR9_MOUSE
|
||||||
| NC score | 0.955880 (rank : 9) | θ value | 4.36332e-79 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z2D0, Q80XL4 | Gene names | Mtmr9, Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 9. | |||||
|
MTMR7_HUMAN
|
||||||
| NC score | 0.951452 (rank : 10) | θ value | 2.38316e-93 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y216, Q68DX4 | Gene names | MTMR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR7_MOUSE
|
||||||
| NC score | 0.950011 (rank : 11) | θ value | 8.47616e-91 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
MTMR3_MOUSE
|
||||||
| NC score | 0.938882 (rank : 12) | θ value | 3.22093e-90 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K296 | Gene names | Mtmr3 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48). | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.834850 (rank : 13) | θ value | 4.96919e-91 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.701921 (rank : 14) | θ value | 5.584e-34 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
MTMRD_HUMAN
|
||||||
| NC score | 0.700854 (rank : 15) | θ value | 1.46948e-34 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86WG5, Q3MJF0, Q68DQ3, Q6P459, Q6PJD1, Q7Z325, Q7Z621, Q86VE2, Q96FE2, Q9C097 | Gene names | SBF2, CMT4B2, KIAA1766, MTMR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myotubularin-related protein 13 (SET-binding factor 2). | |||||
|
DUS7_HUMAN
|
||||||
| NC score | 0.063892 (rank : 16) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
MYCPP_HUMAN
|
||||||
| NC score | 0.059975 (rank : 17) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z401, Q14655, Q86T77, Q8IVX2, Q8NB93 | Gene names | MYCPBP, IRLB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-myc promoter-binding protein. | |||||
|
DUS9_HUMAN
|
||||||
| NC score | 0.050393 (rank : 18) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.050289 (rank : 19) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
DUS6_MOUSE
|
||||||
| NC score | 0.049247 (rank : 20) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS6_HUMAN
|
||||||
| NC score | 0.049212 (rank : 21) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS4_MOUSE
|
||||||
| NC score | 0.046551 (rank : 22) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS4_HUMAN
|
||||||
| NC score | 0.046490 (rank : 23) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS5_HUMAN
|
||||||
| NC score | 0.037605 (rank : 24) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS2_MOUSE
|
||||||
| NC score | 0.037383 (rank : 25) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_HUMAN
|
||||||
| NC score | 0.033928 (rank : 26) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS16_HUMAN
|
||||||
| NC score | 0.032528 (rank : 27) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
SMYD4_MOUSE
|
||||||
| NC score | 0.011810 (rank : 28) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BTK5, Q3U3Z8, Q501N5, Q5SWN4, Q69Z60, Q8BMP3 | Gene names | Smyd4, Kiaa1936 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET and MYND domain-containing protein 4. | |||||
|
PTPRJ_HUMAN
|
||||||
| NC score | 0.011745 (rank : 29) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
PTPRJ_MOUSE
|
||||||
| NC score | 0.011063 (rank : 30) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64455 | Gene names | Ptprj, Byp, Scc1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP beta-like tyrosine phosphatase) (Protein-tyrosine phosphatase receptor type J) (Susceptibility to colon cancer 1). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.009303 (rank : 31) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.008717 (rank : 32) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
AP3M2_HUMAN
|
||||||
| NC score | 0.007078 (rank : 33) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
AP3M2_MOUSE
|
||||||
| NC score | 0.006541 (rank : 34) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.004023 (rank : 35) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
KV1A_HUMAN
|
||||||
| NC score | 0.002267 (rank : 36) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01593 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region AG. | |||||
|
KV1O_HUMAN
|
||||||
| NC score | 0.001159 (rank : 37) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01607 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-I region Rei. | |||||
|
SORL_MOUSE
|
||||||
| NC score | 0.000561 (rank : 38) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||