Please be patient as the page loads
|
HEAT3_MOUSE
|
||||||
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HEAT3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991295 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
HEAT3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 0.279714 (rank : 3) | NC score | 0.023481 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 4) | NC score | 0.080227 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 5) | NC score | 0.080131 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 6) | NC score | 0.081743 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
PKP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 7) | NC score | 0.079105 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
DVL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.034106 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
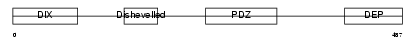
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.034113 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
ARVC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.074819 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
ARVC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.074870 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
MRE11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.033794 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61216, Q62430 | Gene names | Mre11a, Mre11 | |||
|
Domain Architecture |
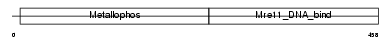
|
|||||
| Description | Double-strand break repair protein MRE11A (MRE11 homolog 1) (MRE11 meiotic recombination 11 homolog A) (MmMRE11A). | |||||
|
PIDD_MOUSE
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.014704 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 2 | |
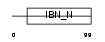
| SwissProt Accessions | Q9ERV7 | Gene names | Lrdd, Pidd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and death domain-containing protein. | |||||
|
ANC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.040200 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJX6, Q5VSG1, Q96DG5, Q96GG4, Q9P2E1 | Gene names | ANAPC2, APC2, KIAA1406 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
EGLX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.023677 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXG6, Q6PAG6, Q8TCJ9, Q8WV55, Q96F22, Q9BW77 | Gene names | PH4 | |||
|
Domain Architecture |
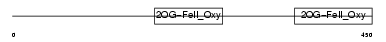
|
|||||
| Description | Putative HIF-prolyl hydroxylase PH-4 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl 4-hydroxylase). | |||||
|
IPO8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.016994 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TMY7, Q811I3, Q8C8A9, Q8R2P6, Q8R3V7 | Gene names | Ipo8 | |||
|
Domain Architecture |
|
|||||
| Description | Importin-8 (Imp8) (Ran-binding protein 8) (RanBP8). | |||||
|
K0226_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.033428 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U62, Q3TAX3, Q3TDF7, Q3TDU2, Q6P9T7, Q6PG18, Q8BMP7, Q8BY22 | Gene names | Kiaa0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
MPIP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.018691 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30305, O43551, Q13971, Q5JX77, Q6RSS1, Q9BRA6 | Gene names | CDC25B, CDC25HU2 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 2 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25B). | |||||
|
SPAG6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.020909 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
ANC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.035482 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZQ7, Q8R2Q1 | Gene names | Anapc2 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
CAND2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.015049 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
K0226_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.031183 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
CHSTA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.012775 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43529 | Gene names | CHST10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST) (huHNK-1ST). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.012463 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
IER2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.022664 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTL4, Q03827, Q2TAZ2 | Gene names | IER2, ETR101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 2 protein (Protein ETR101). | |||||
|
CEP72_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.013991 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D3R3, Q8BM53, Q9D0B7 | Gene names | Cep72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.005738 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
DEN2D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.008535 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VV4, Q3U028, Q9D831 | Gene names | Dennd2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
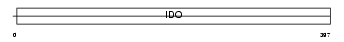
I23O_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.016993 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14902 | Gene names | INDO, IDO | |||
|
Domain Architecture |
|
|||||
| Description | Indoleamine 2,3-dioxygenase (EC 1.13.11.52) (IDO) (Indoleamine-pyrrole 2,3-dioxygenase). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.009517 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
CTND1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.053692 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.053709 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
HEAT3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
HEAT3_HUMAN
|
||||||
| NC score | 0.991295 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.081743 (rank : 3) | θ value | 0.62314 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.080227 (rank : 4) | θ value | 0.47712 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.080131 (rank : 5) | θ value | 0.62314 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
PKP4_HUMAN
|
||||||
| NC score | 0.079105 (rank : 6) | θ value | 0.62314 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
ARVC_MOUSE
|
||||||
| NC score | 0.074870 (rank : 7) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.074819 (rank : 8) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
CTND1_MOUSE
|
||||||
| NC score | 0.053709 (rank : 9) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.053692 (rank : 10) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
ANC2_HUMAN
|
||||||
| NC score | 0.040200 (rank : 11) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJX6, Q5VSG1, Q96DG5, Q96GG4, Q9P2E1 | Gene names | ANAPC2, APC2, KIAA1406 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
ANC2_MOUSE
|
||||||
| NC score | 0.035482 (rank : 12) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZQ7, Q8R2Q1 | Gene names | Anapc2 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 2 (APC2) (Cyclosome subunit 2). | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.034113 (rank : 13) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.034106 (rank : 14) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
MRE11_MOUSE
|
||||||
| NC score | 0.033794 (rank : 15) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61216, Q62430 | Gene names | Mre11a, Mre11 | |||
|
Domain Architecture |

|
|||||
| Description | Double-strand break repair protein MRE11A (MRE11 homolog 1) (MRE11 meiotic recombination 11 homolog A) (MmMRE11A). | |||||
|
K0226_MOUSE
|
||||||
| NC score | 0.033428 (rank : 16) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80U62, Q3TAX3, Q3TDF7, Q3TDU2, Q6P9T7, Q6PG18, Q8BMP7, Q8BY22 | Gene names | Kiaa0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
K0226_HUMAN
|
||||||
| NC score | 0.031183 (rank : 17) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
EGLX_HUMAN
|
||||||
| NC score | 0.023677 (rank : 18) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXG6, Q6PAG6, Q8TCJ9, Q8WV55, Q96F22, Q9BW77 | Gene names | PH4 | |||
|
Domain Architecture |
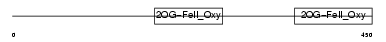
|
|||||
| Description | Putative HIF-prolyl hydroxylase PH-4 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl 4-hydroxylase). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.023481 (rank : 19) | θ value | 0.279714 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
IER2_HUMAN
|
||||||
| NC score | 0.022664 (rank : 20) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTL4, Q03827, Q2TAZ2 | Gene names | IER2, ETR101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 2 protein (Protein ETR101). | |||||
|
SPAG6_MOUSE
|
||||||
| NC score | 0.020909 (rank : 21) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
MPIP2_HUMAN
|
||||||
| NC score | 0.018691 (rank : 22) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30305, O43551, Q13971, Q5JX77, Q6RSS1, Q9BRA6 | Gene names | CDC25B, CDC25HU2 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 2 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25B). | |||||
|
IPO8_MOUSE
|
||||||
| NC score | 0.016994 (rank : 23) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TMY7, Q811I3, Q8C8A9, Q8R2P6, Q8R3V7 | Gene names | Ipo8 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-8 (Imp8) (Ran-binding protein 8) (RanBP8). | |||||
|
I23O_HUMAN
|
||||||
| NC score | 0.016993 (rank : 24) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14902 | Gene names | INDO, IDO | |||
|
Domain Architecture |

|
|||||
| Description | Indoleamine 2,3-dioxygenase (EC 1.13.11.52) (IDO) (Indoleamine-pyrrole 2,3-dioxygenase). | |||||
|
CAND2_MOUSE
|
||||||
| NC score | 0.015049 (rank : 25) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
PIDD_MOUSE
|
||||||
| NC score | 0.014704 (rank : 26) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERV7 | Gene names | Lrdd, Pidd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and death domain-containing protein. | |||||
|
CEP72_MOUSE
|
||||||
| NC score | 0.013991 (rank : 27) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D3R3, Q8BM53, Q9D0B7 | Gene names | Cep72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
CHSTA_HUMAN
|
||||||
| NC score | 0.012775 (rank : 28) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43529 | Gene names | CHST10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST) (huHNK-1ST). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.012463 (rank : 29) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.009517 (rank : 30) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
DEN2D_MOUSE
|
||||||
| NC score | 0.008535 (rank : 31) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VV4, Q3U028, Q9D831 | Gene names | Dennd2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2D. | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.005738 (rank : 32) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||