Please be patient as the page loads
|
DHX29_MOUSE
|
||||||
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DHX29_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991082 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 8.02161e-166 (rank : 3) | NC score | 0.962081 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 3.72618e-163 (rank : 4) | NC score | 0.958497 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 1.56551e-161 (rank : 5) | NC score | 0.958820 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX36_HUMAN
|
||||||
| θ value | 2.26059e-160 (rank : 6) | NC score | 0.965471 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 1.13501e-119 (rank : 7) | NC score | 0.950910 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 8.41738e-115 (rank : 8) | NC score | 0.950356 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
PRP16_HUMAN
|
||||||
| θ value | 4.99229e-75 (rank : 9) | NC score | 0.886012 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 10) | NC score | 0.918826 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX15_HUMAN
|
||||||
| θ value | 1.27544e-62 (rank : 11) | NC score | 0.895407 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| θ value | 1.27544e-62 (rank : 12) | NC score | 0.895340 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 4.84666e-62 (rank : 13) | NC score | 0.917915 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 1.08223e-53 (rank : 14) | NC score | 0.897435 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX8_HUMAN
|
||||||
| θ value | 9.48078e-50 (rank : 15) | NC score | 0.875365 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX35_HUMAN
|
||||||
| θ value | 3.15612e-45 (rank : 16) | NC score | 0.887368 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 17) | NC score | 0.843232 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
DHX34_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 18) | NC score | 0.909736 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX33_MOUSE
|
||||||
| θ value | 2.50074e-42 (rank : 19) | NC score | 0.880864 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX33_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 20) | NC score | 0.876174 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
YTDC2_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 21) | NC score | 0.841624 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 22) | NC score | 0.877153 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DHX40_MOUSE
|
||||||
| θ value | 3.61944e-33 (rank : 23) | NC score | 0.869354 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX40_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 24) | NC score | 0.869695 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 25) | NC score | 0.023961 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 26) | NC score | 0.053711 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 27) | NC score | 0.050987 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.022734 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.006423 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.065469 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.057553 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.062619 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.019307 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.016609 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.031465 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.042779 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.018851 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.003132 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.035860 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.035860 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.017251 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.024007 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.049238 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.033085 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.033088 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.002305 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.022192 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
P52K_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.030649 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
SPA12_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.003090 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TMF5, Q9CQ32 | Gene names | Serpina12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A12 precursor (Visceral adipose-specific serpin) (Visceral adipose tissue-derived serine protease inhibitor) (Vaspin). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.005065 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.032997 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.032997 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.018578 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.014425 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.021316 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.030573 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.032615 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.032620 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
IGB1B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.013267 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ29 | Gene names | Igbp1b | |||
|
Domain Architecture |
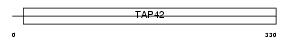
|
|||||
| Description | Immunoglobulin-binding protein 1b (CD79a-binding protein 1b) (Alpha 4- b protein) (Alpha4-b). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.017527 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
3BP5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.014201 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |
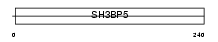
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
ADA30_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.007870 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.009693 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.035472 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.012494 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
IQGA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.007916 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.008523 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.016504 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
RFXK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.002435 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14593, O95839 | Gene names | RFXANK, ANKRA1, RFXB | |||
|
Domain Architecture |
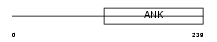
|
|||||
| Description | DNA-binding protein RFXANK (Regulatory factor X subunit B) (RFX-B) (Ankyrin repeat family A protein 1). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.027386 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.017136 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.005174 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CYH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.005158 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
CYH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.005159 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
DPOA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.010318 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14181, Q9BPV3 | Gene names | POLA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase subunit alpha B (DNA polymerase alpha 70 kDa subunit). | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.003155 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
LYAR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.033924 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.012886 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
PRPF3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.023804 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |
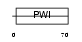
|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
PRPF3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.023542 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.009614 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
STAR7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.009807 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
ABCCB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.002999 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96J66, Q8TDJ0, Q96JA6, Q9BX80 | Gene names | ABCC11, MRP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette transporter sub-family C member 11 (Multidrug resistance-associated protein 8). | |||||
|
CP2CJ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.001349 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33261, P33259, Q8WZB1, Q8WZB2 | Gene names | CYP2C19 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 2C19 (EC 1.14.13.80) ((R)-limonene 6-monooxygenase) (EC 1.14.13.48) ((S)-limonene 6-monooxygenase) (EC 1.14.13.49) ((S)- limonene 7-monooxygenase) (CYPIIC19) (P450-11A) (Mephenytoin 4- hydroxylase) (CYPIIC17) (P450-254C). | |||||
|
DTNA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.008687 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |
|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
FOXJ3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.002594 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein J3. | |||||
|
GGA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.005212 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.000999 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.029091 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.016865 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TNNC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.001123 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
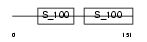
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
VPS53_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.007924 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VIR6, Q8WYW3, Q9BRR2, Q9BY02, Q9NV25 | Gene names | VPS53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
DTNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.007886 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
ITA11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.002755 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKX5, Q8WYI8, Q9UKQ1 | Gene names | ITGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
K0406_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.006413 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43156, Q5JX67, Q96A38, Q9BR47, Q9H4K0 | Gene names | KIAA0406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0406. | |||||
|
MYO3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | -0.001004 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXR4, Q8IX64, Q8IX65, Q8IX66, Q8IX67, Q8IX68 | Gene names | MYO3B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIB (EC 2.7.11.1). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.012354 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NOC2L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.015003 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |
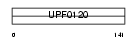
|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.014434 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.007664 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.001191 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
UTP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.011547 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYH9, Q8IX96, Q96BL2, Q9NQ91 | Gene names | UTP6, C17orf40, HCA66, MHAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 6 homolog (Multiple hat domains protein) (Hepatocellular carcinoma-associated antigen 66). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.025508 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.991082 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX36_HUMAN
|
||||||
| NC score | 0.965471 (rank : 3) | θ value | 2.26059e-160 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.962081 (rank : 4) | θ value | 8.02161e-166 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.958820 (rank : 5) | θ value | 1.56551e-161 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.958497 (rank : 6) | θ value | 3.72618e-163 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.950910 (rank : 7) | θ value | 1.13501e-119 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.950356 (rank : 8) | θ value | 8.41738e-115 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.918826 (rank : 9) | θ value | 1.50663e-63 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.917915 (rank : 10) | θ value | 4.84666e-62 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX34_HUMAN
|
||||||
| NC score | 0.909736 (rank : 11) | θ value | 2.95404e-43 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.897435 (rank : 12) | θ value | 1.08223e-53 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX15_HUMAN
|
||||||
| NC score | 0.895407 (rank : 13) | θ value | 1.27544e-62 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| NC score | 0.895340 (rank : 14) | θ value | 1.27544e-62 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX35_HUMAN
|
||||||
| NC score | 0.887368 (rank : 15) | θ value | 3.15612e-45 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
PRP16_HUMAN
|
||||||
| NC score | 0.886012 (rank : 16) | θ value | 4.99229e-75 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX33_MOUSE
|
||||||
| NC score | 0.880864 (rank : 17) | θ value | 2.50074e-42 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.877153 (rank : 18) | θ value | 6.59618e-35 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DHX33_HUMAN
|
||||||
| NC score | 0.876174 (rank : 19) | θ value | 4.71619e-41 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX8_HUMAN
|
||||||
| NC score | 0.875365 (rank : 20) | θ value | 9.48078e-50 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX40_HUMAN
|
||||||
| NC score | 0.869695 (rank : 21) | θ value | 8.06329e-33 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| NC score | 0.869354 (rank : 22) | θ value | 3.61944e-33 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.843232 (rank : 23) | θ value | 1.32601e-43 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
YTDC2_HUMAN
|
||||||
| NC score | 0.841624 (rank : 24) | θ value | 2.96089e-35 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.065469 (rank : 25) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.062619 (rank : 26) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.057553 (rank : 27) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.053711 (rank : 28) | θ value | 0.0193708 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.050987 (rank : 29) | θ value | 0.0252991 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.049238 (rank : 30) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.042779 (rank : 31) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.035860 (rank : 32) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.035860 (rank : 33) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.035472 (rank : 34) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
LYAR_HUMAN
|
||||||
| NC score | 0.033924 (rank : 35) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NX58, Q6FI78, Q9NYS1 | Gene names | LYAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.033088 (rank : 36) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.033085 (rank : 37) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.032997 (rank : 38) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.032997 (rank : 39) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.032620 (rank : 40) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.032615 (rank : 41) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.031465 (rank : 42) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.030649 (rank : 43) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.030573 (rank : 44) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.029091 (rank : 45) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.027386 (rank : 46) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.025508 (rank : 47) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.024007 (rank : 48) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.023961 (rank : 49) | θ value | 0.0113563 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
PRPF3_HUMAN
|
||||||
| NC score | 0.023804 (rank : 50) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
PRPF3_MOUSE
|
||||||
| NC score | 0.023542 (rank : 51) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.022734 (rank : 52) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.022192 (rank : 53) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.021316 (rank : 54) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.019307 (rank : 55) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.018851 (rank : 56) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.018578 (rank : 57) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.017527 (rank : 58) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.017251 (rank : 59) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.017136 (rank : 60) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.016865 (rank : 61) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.016609 (rank : 62) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.016504 (rank : 63) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
NOC2L_MOUSE
|
||||||
| NC score | 0.015003 (rank : 64) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.014434 (rank : 65) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.014425 (rank : 66) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
3BP5_HUMAN
|
||||||
| NC score | 0.014201 (rank : 67) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
IGB1B_MOUSE
|
||||||
| NC score | 0.013267 (rank : 68) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ29 | Gene names | Igbp1b | |||
|
Domain Architecture |

|
|||||
| Description | Immunoglobulin-binding protein 1b (CD79a-binding protein 1b) (Alpha 4- b protein) (Alpha4-b). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.012886 (rank : 69) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.012494 (rank : 70) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.012354 (rank : 71) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
UTP6_HUMAN
|
||||||
| NC score | 0.011547 (rank : 72) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYH9, Q8IX96, Q96BL2, Q9NQ91 | Gene names | UTP6, C17orf40, HCA66, MHAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 6 homolog (Multiple hat domains protein) (Hepatocellular carcinoma-associated antigen 66). | |||||
|
DPOA2_HUMAN
|
||||||
| NC score | 0.010318 (rank : 73) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14181, Q9BPV3 | Gene names | POLA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase subunit alpha B (DNA polymerase alpha 70 kDa subunit). | |||||
|
STAR7_MOUSE
|
||||||
| NC score | 0.009807 (rank : 74) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.009693 (rank : 75) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.009614 (rank : 76) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
DTNA_MOUSE
|
||||||
| NC score | 0.008687 (rank : 77) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.008523 (rank : 78) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
VPS53_HUMAN
|
||||||
| NC score | 0.007924 (rank : 79) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VIR6, Q8WYW3, Q9BRR2, Q9BY02, Q9NV25 | Gene names | VPS53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
IQGA3_HUMAN
|
||||||
| NC score | 0.007916 (rank : 80) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86VI3 | Gene names | IQGAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP3. | |||||
|
DTNA_HUMAN
|
||||||
| NC score | 0.007886 (rank : 81) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
ADA30_HUMAN
|
||||||
| NC score | 0.007870 (rank : 82) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.007664 (rank : 83) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.006423 (rank : 84) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
K0406_HUMAN
|
||||||
| NC score | 0.006413 (rank : 85) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43156, Q5JX67, Q96A38, Q9BR47, Q9H4K0 | Gene names | KIAA0406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0406. | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.005212 (rank : 86) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.005174 (rank : 87) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CYH2_MOUSE
|
||||||
| NC score | 0.005159 (rank : 88) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63034, O89099, P97695, Q3UP39 | Gene names | Pscd2, Sec7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytohesin-2 (CLM2) (SEC7 homolog B) (msec7-2). | |||||
|
CYH2_HUMAN
|
||||||
| NC score | 0.005158 (rank : 89) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99418, Q92958 | Gene names | PSCD2, ARNO | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-2 (ARF nucleotide-binding site opener) (ARNO protein) (ARF exchange factor). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.005065 (rank : 90) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.003155 (rank : 91) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.003132 (rank : 92) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
SPA12_MOUSE
|
||||||
| NC score | 0.003090 (rank : 93) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TMF5, Q9CQ32 | Gene names | Serpina12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A12 precursor (Visceral adipose-specific serpin) (Visceral adipose tissue-derived serine protease inhibitor) (Vaspin). | |||||
|
ABCCB_HUMAN
|
||||||
| NC score | 0.002999 (rank : 94) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96J66, Q8TDJ0, Q96JA6, Q9BX80 | Gene names | ABCC11, MRP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette transporter sub-family C member 11 (Multidrug resistance-associated protein 8). | |||||
|
ITA11_HUMAN
|
||||||
| NC score | 0.002755 (rank : 95) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKX5, Q8WYI8, Q9UKQ1 | Gene names | ITGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
FOXJ3_MOUSE
|
||||||
| NC score | 0.002594 (rank : 96) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BUR3, Q3UTD8 | Gene names | Foxj3, Kiaa1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
RFXK_HUMAN
|
||||||
| NC score | 0.002435 (rank : 97) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14593, O95839 | Gene names | RFXANK, ANKRA1, RFXB | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFXANK (Regulatory factor X subunit B) (RFX-B) (Ankyrin repeat family A protein 1). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.002305 (rank : 98) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
CP2CJ_HUMAN
|
||||||
| NC score | 0.001349 (rank : 99) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33261, P33259, Q8WZB1, Q8WZB2 | Gene names | CYP2C19 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C19 (EC 1.14.13.80) ((R)-limonene 6-monooxygenase) (EC 1.14.13.48) ((S)-limonene 6-monooxygenase) (EC 1.14.13.49) ((S)- limonene 7-monooxygenase) (CYPIIC19) (P450-11A) (Mephenytoin 4- hydroxylase) (CYPIIC17) (P450-254C). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.001191 (rank : 100) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.001123 (rank : 101) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.000999 (rank : 102) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
MYO3B_HUMAN
|
||||||
| NC score | -0.001004 (rank : 103) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXR4, Q8IX64, Q8IX65, Q8IX66, Q8IX67, Q8IX68 | Gene names | MYO3B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIB (EC 2.7.11.1). | |||||