Please be patient as the page loads
|
NF2L2_HUMAN
|
||||||
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NF2L2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.958956 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NF2L1_HUMAN
|
||||||
| θ value | 1.36269e-64 (rank : 3) | NC score | 0.874619 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| θ value | 2.25659e-51 (rank : 4) | NC score | 0.858907 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
NF2L3_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 5) | NC score | 0.839513 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NFE2_HUMAN
|
||||||
| θ value | 6.81017e-40 (rank : 6) | NC score | 0.902181 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
NF2L3_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 7) | NC score | 0.845045 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
BACH2_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 8) | NC score | 0.378538 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH2_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 9) | NC score | 0.381660 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH1_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 10) | NC score | 0.380064 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
BACH1_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 11) | NC score | 0.381117 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
JUND_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 12) | NC score | 0.332501 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.335471 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUN_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 14) | NC score | 0.327839 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUN_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 15) | NC score | 0.328212 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUNB_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 16) | NC score | 0.329107 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUNB_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 17) | NC score | 0.324241 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor jun-B. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 18) | NC score | 0.048315 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 19) | NC score | 0.068435 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MAF_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.171022 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.015863 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.024753 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.067594 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MAF_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.168054 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
3MG_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.059261 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q04841, Q64182 | Gene names | Mpg, Mid1 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-3-methyladenine glycosylase (EC 3.2.2.21) (3-methyladenine DNA glycosidase) (ADPG) (3-alkyladenine DNA glycosylase) (N-methylpurine- DNA glycosylase). | |||||
|
GR65_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.038794 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
ATF6A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.094944 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.019674 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
BATF_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.175558 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |
|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.022972 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.010796 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.025267 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.032885 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.034618 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
SPB4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.008840 (rank : 123) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48594 | Gene names | SERPINB4, SCCA2 | |||
|
Domain Architecture |
|
|||||
| Description | Serpin B4 (Squamous cell carcinoma antigen 2) (SCCA-2) (Leupin). | |||||
|
TACC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.024177 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.005763 (rank : 128) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
FOS_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.120061 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOS_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.121434 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
MAFB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.174394 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
MAFB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.174824 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
N4BP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.041537 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.018908 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.019635 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.004518 (rank : 130) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
KCC2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.002384 (rank : 133) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13554, O95437, O95438, O95599, Q9UGH7, Q9UGH8, Q9UGH9, Q9UNX0, Q9UNX7, Q9UP00, Q9Y5N4, Q9Y6F4 | Gene names | CAMK2B, CAM2, CAMKB | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II beta chain (EC 2.7.11.17) (CaM-kinase II beta chain) (CaM kinase II subunit beta) (CaMK-II subunit beta). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.017048 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
SAPS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.018379 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.016882 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.019593 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.008286 (rank : 125) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.031498 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.019935 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
PKN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.003924 (rank : 132) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16513, Q9H1W4 | Gene names | PKN2, PRK2, PRKCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase N2 (EC 2.7.11.13) (Protein kinase C- like 2) (Protein-kinase C-related kinase 2). | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.026898 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.022004 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CKAP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.025967 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.014969 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.033153 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
PKN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.004430 (rank : 131) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWW9, Q80WS2, Q8BJL7 | Gene names | Pkn2, Prkcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N2 (EC 2.7.11.13) (Protein kinase C- like 2) (Protein-kinase C-related kinase 2). | |||||
|
SUFU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.031944 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMX1, Q7LCP7, Q9NT90, Q9NZ07, Q9UHK2, Q9UHM8, Q9UMY0 | Gene names | SUFU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of fused homolog (SUFUH). | |||||
|
SUFU_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.032067 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0P7, Q8C8B4, Q99JG0, Q9D521, Q9JLU1 | Gene names | Sufu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of fused homolog. | |||||
|
3BP5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.019325 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |
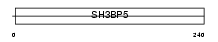
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
ATF4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.022758 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
CCD45_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.018324 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
CLMN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.008472 (rank : 124) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CREB3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.158798 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
DCR1C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.016415 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4J0, Q8BG72, Q8BTT1 | Gene names | Dclre1c, Art, Snm1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemis protein (EC 3.1.-.-) (DNA cross-link repair 1C protein) (mArt protein) (SNM1-like protein). | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.019892 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.018632 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.024756 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
UBN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.026497 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.019767 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CASL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.009830 (rank : 121) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |
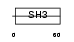
|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.015729 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.012886 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.018025 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.028584 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
M3K3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.002028 (rank : 135) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99759 | Gene names | MAP3K3, MAPKKK3, MEKK3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.021115 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.021088 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SMAD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.009576 (rank : 122) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SPA9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.005372 (rank : 129) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86WD7, Q6UWP9, Q86WD6, Q86YP6, Q86YP7 | Gene names | SERPINA9, GCET1, SERPINA11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A9 precursor (Germinal center B-cell expressed transcript 1 protein). | |||||
|
ZBED4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.019140 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75132, Q9UGG8 | Gene names | ZBED4, KIAA0637 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 4. | |||||
|
AT2B2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.007015 (rank : 126) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.006732 (rank : 127) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
BANK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.014921 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80VH0, Q3U178, Q8BRV6, Q8BRY8 | Gene names | Bank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats (Protein AVIEF). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.013246 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.020020 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.014576 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.009863 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.015355 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.012247 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
PTN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.002174 (rank : 134) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
SDC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.015283 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43407 | Gene names | Sdc2, Hspg1, Synd2 | |||
|
Domain Architecture |
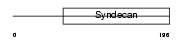
|
|||||
| Description | Syndecan-2 precursor (SYND2) (Fibroglycan) (Heparan sulfate proteoglycan core protein) (HSPG). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.020324 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
AKAP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.017126 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.014977 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.021099 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.010178 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
EI2BE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.010270 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13144, Q96D04 | Gene names | EIF2B5, EIF2BE | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit epsilon (eIF-2B GDP-GTP exchange factor subunit epsilon). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.012642 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.012743 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.123780 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.123081 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.013546 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
K1024_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.010757 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3V7, Q69ZS9 | Gene names | Kiaa1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024 (Protein DD1). | |||||
|
M3K3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.001607 (rank : 136) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61084 | Gene names | Map3k3, Mekk3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||
|
SIN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.016622 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BPZ7, Q00426 | Gene names | MAPKAP1, SIN1 | |||
|
Domain Architecture |
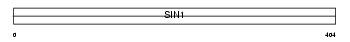
|
|||||
| Description | Stress-activated map kinase-interacting protein 1 (SAPK-interacting protein 1) (Putative Ras inhibitor JC310). | |||||
|
ATF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.054790 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.051839 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
ATF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.058128 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.058225 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.144150 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
ATF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.148653 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.062287 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.058053 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
BATF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.149154 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |
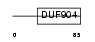
|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
CREB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.068353 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.091378 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
CREM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.063083 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |
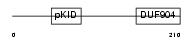
|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.063788 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
FOSB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.112893 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.110809 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
FOSL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.114518 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.109921 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
MAFF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.081906 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULX9, Q9Y525 | Gene names | MAFF | |||
|
Domain Architecture |
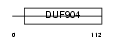
|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F) (U-Maf). | |||||
|
MAFF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.082726 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54791 | Gene names | Maff | |||
|
Domain Architecture |
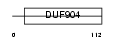
|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F). | |||||
|
MAFG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.135730 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15525 | Gene names | MAFG | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G) (hMAF). | |||||
|
MAFG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.135622 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
MAFK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.148805 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAFK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.145717 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
NRL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.081348 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
Domain Architecture |
|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
NRL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.092909 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P54846 | Gene names | Nrl | |||
|
Domain Architecture |
|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL). | |||||
|
XBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.183683 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
XBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.160090 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.958956 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NFE2_HUMAN
|
||||||
| NC score | 0.902181 (rank : 3) | θ value | 6.81017e-40 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
NF2L1_HUMAN
|
||||||
| NC score | 0.874619 (rank : 4) | θ value | 1.36269e-64 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| NC score | 0.858907 (rank : 5) | θ value | 2.25659e-51 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
NF2L3_MOUSE
|
||||||
| NC score | 0.845045 (rank : 6) | θ value | 7.52953e-39 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.839513 (rank : 7) | θ value | 2.11701e-41 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
BACH2_MOUSE
|
||||||
| NC score | 0.381660 (rank : 8) | θ value | 8.95645e-16 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH1_HUMAN
|
||||||
| NC score | 0.381117 (rank : 9) | θ value | 9.90251e-15 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| NC score | 0.380064 (rank : 10) | θ value | 7.58209e-15 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
BACH2_HUMAN
|
||||||
| NC score | 0.378538 (rank : 11) | θ value | 6.85773e-16 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.335471 (rank : 12) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_HUMAN
|
||||||
| NC score | 0.332501 (rank : 13) | θ value | 0.00102713 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUNB_HUMAN
|
||||||
| NC score | 0.329107 (rank : 14) | θ value | 0.00298849 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUN_MOUSE
|
||||||
| NC score | 0.328212 (rank : 15) | θ value | 0.00134147 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUN_HUMAN
|
||||||
| NC score | 0.327839 (rank : 16) | θ value | 0.00134147 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUNB_MOUSE
|
||||||
| NC score | 0.324241 (rank : 17) | θ value | 0.00298849 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
XBP1_HUMAN
|
||||||
| NC score | 0.183683 (rank : 18) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
BATF_MOUSE
|
||||||
| NC score | 0.175558 (rank : 19) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
MAFB_MOUSE
|
||||||
| NC score | 0.174824 (rank : 20) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MAFB_HUMAN
|
||||||
| NC score | 0.174394 (rank : 21) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
MAF_HUMAN
|
||||||
| NC score | 0.171022 (rank : 22) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAF_MOUSE
|
||||||
| NC score | 0.168054 (rank : 23) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
XBP1_MOUSE
|
||||||
| NC score | 0.160090 (rank : 24) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
CREB3_MOUSE
|
||||||
| NC score | 0.158798 (rank : 25) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
BATF_HUMAN
|
||||||
| NC score | 0.149154 (rank : 26) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
MAFK_HUMAN
|
||||||
| NC score | 0.148805 (rank : 27) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
ATF3_MOUSE
|
||||||
| NC score | 0.148653 (rank : 28) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
MAFK_MOUSE
|
||||||
| NC score | 0.145717 (rank : 29) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
ATF3_HUMAN
|
||||||
| NC score | 0.144150 (rank : 30) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
MAFG_HUMAN
|
||||||
| NC score | 0.135730 (rank : 31) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15525 | Gene names | MAFG | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G) (hMAF). | |||||
|
MAFG_MOUSE
|
||||||
| NC score | 0.135622 (rank : 32) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.123780 (rank : 33) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.123081 (rank : 34) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOS_MOUSE
|
||||||
| NC score | 0.121434 (rank : 35) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.120061 (rank : 36) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOSL2_HUMAN
|
||||||
| NC score | 0.114518 (rank : 37) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.112893 (rank : 38) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.110809 (rank : 39) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
FOSL2_MOUSE
|
||||||
| NC score | 0.109921 (rank : 40) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.094944 (rank : 41) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
NRL_MOUSE
|
||||||
| NC score | 0.092909 (rank : 42) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P54846 | Gene names | Nrl | |||
|
Domain Architecture |

|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.091378 (rank : 43) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
MAFF_MOUSE
|
||||||
| NC score | 0.082726 (rank : 44) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O54791 | Gene names | Maff | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F). | |||||
|
MAFF_HUMAN
|
||||||
| NC score | 0.081906 (rank : 45) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ULX9, Q9Y525 | Gene names | MAFF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F) (U-Maf). | |||||
|
NRL_HUMAN
|
||||||
| NC score | 0.081348 (rank : 46) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
Domain Architecture |

|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.068435 (rank : 47) | θ value | 0.0148317 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CREB3_HUMAN
|
||||||
| NC score | 0.068353 (rank : 48) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.067594 (rank : 49) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CREM_MOUSE
|
||||||
| NC score | 0.063788 (rank : 50) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_HUMAN
|
||||||
| NC score | 0.063083 (rank : 51) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.062287 (rank : 52) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
3MG_MOUSE
|
||||||
| NC score | 0.059261 (rank : 53) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q04841, Q64182 | Gene names | Mpg, Mid1 | |||
|
Domain Architecture |
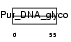
|
|||||
| Description | DNA-3-methyladenine glycosylase (EC 3.2.2.21) (3-methyladenine DNA glycosidase) (ADPG) (3-alkyladenine DNA glycosylase) (N-methylpurine- DNA glycosylase). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.058225 (rank : 54) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.058128 (rank : 55) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.058053 (rank : 56) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF1_HUMAN
|
||||||
| NC score | 0.054790 (rank : 57) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF1_MOUSE
|
||||||
| NC score | 0.051839 (rank : 58) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.048315 (rank : 59) | θ value | 0.00869519 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
N4BP2_HUMAN
|
||||||
| NC score | 0.041537 (rank : 60) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.038794 (rank : 61) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.034618 (rank : 62) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.033153 (rank : 63) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.032885 (rank : 64) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
SUFU_MOUSE
|
||||||
| NC score | 0.032067 (rank : 65) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0P7, Q8C8B4, Q99JG0, Q9D521, Q9JLU1 | Gene names | Sufu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of fused homolog. | |||||
|
SUFU_HUMAN
|
||||||
| NC score | 0.031944 (rank : 66) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UMX1, Q7LCP7, Q9NT90, Q9NZ07, Q9UHK2, Q9UHM8, Q9UMY0 | Gene names | SUFU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of fused homolog (SUFUH). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.031498 (rank : 67) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.028584 (rank : 68) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.026898 (rank : 69) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
UBN1_HUMAN
|
||||||
| NC score | 0.026497 (rank : 70) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
CKAP4_MOUSE
|
||||||
| NC score | 0.025967 (rank : 71) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.025267 (rank : 72) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.024756 (rank : 73) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.024753 (rank : 74) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TACC1_HUMAN
|
||||||
| NC score | 0.024177 (rank : 75) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.022972 (rank : 76) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
ATF4_MOUSE
|
||||||
| NC score | 0.022758 (rank : 77) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.022004 (rank : 78) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.021115 (rank : 79) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.021099 (rank : 80) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.021088 (rank : 81) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.020324 (rank : 82) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.020020 (rank : 83) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.019935 (rank : 84) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.019892 (rank : 85) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.019767 (rank : 86) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.019674 (rank : 87) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.019635 (rank : 88) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.019593 (rank : 89) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
3BP5_HUMAN
|
||||||
| NC score | 0.019325 (rank : 90) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
ZBED4_HUMAN
|
||||||
| NC score | 0.019140 (rank : 91) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75132, Q9UGG8 | Gene names | ZBED4, KIAA0637 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 4. | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.018908 (rank : 92) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.018632 (rank : 93) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
SAPS3_MOUSE
|
||||||
| NC score | 0.018379 (rank : 94) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
CCD45_MOUSE
|
||||||
| NC score | 0.018324 (rank : 95) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BVV7 | Gene names | Ccdc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 45. | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.018025 (rank : 96) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
AKAP8_HUMAN
|
||||||
| NC score | 0.017126 (rank : 97) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.017048 (rank : 98) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.016882 (rank : 99) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
SIN1_HUMAN
|
||||||
| NC score | 0.016622 (rank : 100) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BPZ7, Q00426 | Gene names | MAPKAP1, SIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-activated map kinase-interacting protein 1 (SAPK-interacting protein 1) (Putative Ras inhibitor JC310). | |||||
|
DCR1C_MOUSE
|
||||||
| NC score | 0.016415 (rank : 101) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4J0, Q8BG72, Q8BTT1 | Gene names | Dclre1c, Art, Snm1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Artemis protein (EC 3.1.-.-) (DNA cross-link repair 1C protein) (mArt protein) (SNM1-like protein). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.015863 (rank : 102) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.015729 (rank : 103) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.015355 (rank : 104) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
SDC2_MOUSE
|
||||||
| NC score | 0.015283 (rank : 105) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43407 | Gene names | Sdc2, Hspg1, Synd2 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-2 precursor (SYND2) (Fibroglycan) (Heparan sulfate proteoglycan core protein) (HSPG). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.014977 (rank : 106) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.014969 (rank : 107) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
BANK1_MOUSE
|
||||||
| NC score | 0.014921 (rank : 108) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80VH0, Q3U178, Q8BRV6, Q8BRY8 | Gene names | Bank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats (Protein AVIEF). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.014576 (rank : 109) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.013546 (rank : 110) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.013246 (rank : 111) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.012886 (rank : 112) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.012743 (rank : 113) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.012642 (rank : 114) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.012247 (rank : 115) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.010796 (rank : 116) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
K1024_MOUSE
|
||||||
| NC score | 0.010757 (rank : 117) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3V7, Q69ZS9 | Gene names | Kiaa1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024 (Protein DD1). | |||||
|
EI2BE_HUMAN
|
||||||
| NC score | 0.010270 (rank : 118) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13144, Q96D04 | Gene names | EIF2B5, EIF2BE | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit epsilon (eIF-2B GDP-GTP exchange factor subunit epsilon). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.010178 (rank : 119) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.009863 (rank : 120) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
CASL_HUMAN
|
||||||
| NC score | 0.009830 (rank : 121) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
SMAD4_HUMAN
|
||||||
| NC score | 0.009576 (rank : 122) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SPB4_HUMAN
|
||||||
| NC score | 0.008840 (rank : 123) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48594 | Gene names | SERPINB4, SCCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B4 (Squamous cell carcinoma antigen 2) (SCCA-2) (Leupin). | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.008472 (rank : 124) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.008286 (rank : 125) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
AT2B2_HUMAN
|
||||||
| NC score | 0.007015 (rank : 126) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01814, O00766, Q12994, Q16818 | Gene names | ATP2B2, PMCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
AT2B2_MOUSE
|
||||||
| NC score | 0.006732 (rank : 127) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0K7, O88863 | Gene names | Atp2b2, Pmca2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 2 (EC 3.6.3.8) (PMCA2) (Plasma membrane calcium pump isoform 2) (Plasma membrane calcium ATPase isoform 2). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.005763 (rank : 128) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
SPA9_HUMAN
|
||||||
| NC score | 0.005372 (rank : 129) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86WD7, Q6UWP9, Q86WD6, Q86YP6, Q86YP7 | Gene names | SERPINA9, GCET1, SERPINA11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A9 precursor (Germinal center B-cell expressed transcript 1 protein). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.004518 (rank : 130) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
PKN2_MOUSE
|
||||||
| NC score | 0.004430 (rank : 131) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWW9, Q80WS2, Q8BJL7 | Gene names | Pkn2, Prkcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N2 (EC 2.7.11.13) (Protein kinase C- like 2) (Protein-kinase C-related kinase 2). | |||||
|
PKN2_HUMAN
|
||||||
| NC score | 0.003924 (rank : 132) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16513, Q9H1W4 | Gene names | PKN2, PRK2, PRKCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase N2 (EC 2.7.11.13) (Protein kinase C- like 2) (Protein-kinase C-related kinase 2). | |||||
|
KCC2B_HUMAN
|
||||||
| NC score | 0.002384 (rank : 133) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13554, O95437, O95438, O95599, Q9UGH7, Q9UGH8, Q9UGH9, Q9UNX0, Q9UNX7, Q9UP00, Q9Y5N4, Q9Y6F4 | Gene names | CAMK2B, CAM2, CAMKB | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II beta chain (EC 2.7.11.17) (CaM-kinase II beta chain) (CaM kinase II subunit beta) (CaMK-II subunit beta). | |||||
|
PTN3_HUMAN
|
||||||
| NC score | 0.002174 (rank : 134) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26045 | Gene names | PTPN3, PTPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase H1) (PTP-H1). | |||||
|
M3K3_HUMAN
|
||||||
| NC score | 0.002028 (rank : 135) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 885 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99759 | Gene names | MAP3K3, MAPKKK3, MEKK3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||
|
M3K3_MOUSE
|
||||||
| NC score | 0.001607 (rank : 136) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61084 | Gene names | Map3k3, Mekk3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||