Please be patient as the page loads
|
NF2L3_HUMAN
|
||||||
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NF2L3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 150 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NF2L3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.971962 (rank : 2) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NF2L1_MOUSE
|
||||||
| θ value | 1.95861e-79 (rank : 3) | NC score | 0.888060 (rank : 4) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
NF2L1_HUMAN
|
||||||
| θ value | 2.47765e-74 (rank : 4) | NC score | 0.890189 (rank : 3) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 5.95217e-44 (rank : 5) | NC score | 0.843105 (rank : 6) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 6) | NC score | 0.839513 (rank : 7) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
NFE2_HUMAN
|
||||||
| θ value | 1.6774e-38 (rank : 7) | NC score | 0.885162 (rank : 5) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
BACH2_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 8) | NC score | 0.381698 (rank : 10) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH1_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 9) | NC score | 0.389125 (rank : 8) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 10) | NC score | 0.388202 (rank : 9) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
BACH2_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 11) | NC score | 0.378026 (rank : 11) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
MAFB_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 12) | NC score | 0.257415 (rank : 18) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
JUNB_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.335781 (rank : 17) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUN_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.343621 (rank : 15) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUN_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 15) | NC score | 0.344231 (rank : 14) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
MAFB_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 16) | NC score | 0.256038 (rank : 19) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
JUNB_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 17) | NC score | 0.338499 (rank : 16) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUND_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 18) | NC score | 0.348617 (rank : 12) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 19) | NC score | 0.346206 (rank : 13) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
MAFK_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.223682 (rank : 21) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAFK_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.226192 (rank : 20) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.054657 (rank : 69) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.040901 (rank : 78) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.045165 (rank : 72) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.043856 (rank : 74) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
IRX2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.025455 (rank : 107) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81066, O55121, Q9ERN1 | Gene names | Irx2, Irxa2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.027432 (rank : 104) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.022894 (rank : 115) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.011923 (rank : 138) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
SIAL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.060115 (rank : 64) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |
|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.008290 (rank : 151) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
FOS_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.169227 (rank : 36) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOS_MOUSE
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.170296 (rank : 35) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
MAF_HUMAN
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.217604 (rank : 23) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 35) | NC score | 0.013558 (rank : 135) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
CREB3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.184674 (rank : 31) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
FOSL2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.168259 (rank : 37) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 38) | NC score | 0.166948 (rank : 38) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
MAF_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.219538 (rank : 22) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
NCOAT_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.039640 (rank : 79) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
S30BP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.064546 (rank : 59) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHR5, Q8N1R5, Q8TDR8, Q96D27 | Gene names | SAP30BP, HCNGP, HTRG, HTRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAP30-binding protein (Transcriptional regulator protein HCNGP). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.043683 (rank : 75) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
XBP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.203274 (rank : 25) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.011182 (rank : 141) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
XBP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.217316 (rank : 24) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.163336 (rank : 39) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.162399 (rank : 40) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
LYAR_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.038320 (rank : 83) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.034176 (rank : 89) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.032984 (rank : 95) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
PKP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.020781 (rank : 120) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.009669 (rank : 147) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
RPO3G_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.054767 (rank : 68) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15318 | Gene names | POLR3G, RPC32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.045750 (rank : 71) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.024741 (rank : 109) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.025591 (rank : 105) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
KI20A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.021970 (rank : 116) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95235 | Gene names | KIF20A, RAB6KIFL | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin family member 20A (Rabkinesin-6) (Rab6-interacting kinesin- like protein) (GG10_2). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.036820 (rank : 85) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.038425 (rank : 81) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.034371 (rank : 88) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
S30BP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.060123 (rank : 63) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02614, Q8VDJ5 | Gene names | Sap30bp, Hcngp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAP30-binding protein (Transcriptional regulator protein HCNGP). | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.028868 (rank : 102) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.021671 (rank : 117) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.033327 (rank : 94) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.069704 (rank : 57) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.016516 (rank : 127) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
PP2CG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.025099 (rank : 108) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
SEMG1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.039474 (rank : 80) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
TFE2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.023110 (rank : 114) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |
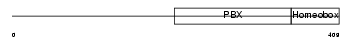
|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
TXD13_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.032518 (rank : 96) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |
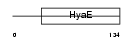
|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.033510 (rank : 92) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.042747 (rank : 76) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.023747 (rank : 112) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.172621 (rank : 33) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.170589 (rank : 34) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
MAFG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.198395 (rank : 28) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15525 | Gene names | MAFG | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G) (hMAF). | |||||
|
MAFG_MOUSE
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.199572 (rank : 27) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.033585 (rank : 90) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.027796 (rank : 103) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PTMS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.063730 (rank : 60) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.006267 (rank : 160) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.025526 (rank : 106) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ARS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.033355 (rank : 93) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.023989 (rank : 111) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.070055 (rank : 56) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MA1A2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.014470 (rank : 133) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P39098, Q5SUY6, Q5SUY7, Q60599, Q9CUY9 | Gene names | Man1a2, Man1b | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.024240 (rank : 110) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PKN2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.002810 (rank : 163) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BWW9, Q80WS2, Q8BJL7 | Gene names | Pkn2, Prkcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N2 (EC 2.7.11.13) (Protein kinase C- like 2) (Protein-kinase C-related kinase 2). | |||||
|
PTMA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.067746 (rank : 58) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.044153 (rank : 73) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
ATF3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.184922 (rank : 30) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
ATF3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.190239 (rank : 29) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
CR025_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.029669 (rank : 100) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96B23, Q5XG78, Q6N058, Q86TB5, Q8TCQ5 | Gene names | C18orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25. | |||||
|
NFM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.008214 (rank : 153) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.032398 (rank : 98) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
WDR43_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.020075 (rank : 121) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
BATF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.201524 (rank : 26) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |
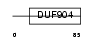
|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.032451 (rank : 97) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
DACT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.019831 (rank : 122) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.031000 (rank : 99) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.054773 (rank : 67) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.016132 (rank : 128) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |
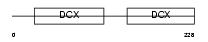
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
SMCE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.018136 (rank : 124) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
ATF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.074327 (rank : 50) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.071545 (rank : 55) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
BATF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.183988 (rank : 32) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |
|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.009563 (rank : 148) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
FOXI1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.004909 (rank : 161) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922I5, Q5SRI5, Q9D299 | Gene names | Foxi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I1. | |||||
|
GET1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.021480 (rank : 118) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95210 | Gene names | ames=GENX-3414 | |||
|
Domain Architecture |

|
|||||
| Description | Genethonin-1. | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.013973 (rank : 134) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.029649 (rank : 101) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
NRL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.148865 (rank : 41) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54846 | Gene names | Nrl | |||
|
Domain Architecture |
|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL). | |||||
|
TNNT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.015956 (rank : 130) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
UTP20_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.021063 (rank : 119) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.010815 (rank : 143) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
CACB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.009437 (rank : 149) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CC27, Q8C5J5, Q9CTQ6 | Gene names | Cacnb2, Cacnlb2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2). | |||||
|
CSTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.006406 (rank : 159) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4D0, Q9BSS0 | Gene names | CLSTN2, CS2 | |||
|
Domain Architecture |
|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.016126 (rank : 129) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
KI21B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.010119 (rank : 145) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
NDUA7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.023353 (rank : 113) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1P6 | Gene names | Ndufa7 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase subunit B14.5a) (Complex I-B14.5a) (CI-B14.5a). | |||||
|
SRPK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.009843 (rank : 146) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O54781, Q8VCD9 | Gene names | Srpk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.008191 (rank : 154) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.012189 (rank : 137) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.011230 (rank : 140) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CREB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.060749 (rank : 62) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.060769 (rank : 61) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |
|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.059346 (rank : 65) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.033572 (rank : 91) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
NRL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.137506 (rank : 42) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
Domain Architecture |
|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.007623 (rank : 156) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
R3HD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.013275 (rank : 136) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.010684 (rank : 144) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
TXD11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.011779 (rank : 139) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.007509 (rank : 157) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.018034 (rank : 125) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.018297 (rank : 123) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.035524 (rank : 86) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
HDGF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.016930 (rank : 126) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.038376 (rank : 82) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.007855 (rank : 155) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.004873 (rank : 162) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.008257 (rank : 152) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.042344 (rank : 77) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
RNF13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.006616 (rank : 158) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54965, O54966 | Gene names | Rnf13, Rzf | |||
|
Domain Architecture |
|
|||||
| Description | RING finger protein 13. | |||||
|
RPO3G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.035171 (rank : 87) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NXY9, Q8K0W5, Q9CV05 | Gene names | Polr3g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
SG16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.036975 (rank : 84) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
SPAT7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.011010 (rank : 142) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0W8, Q8WX30, Q96HF3, Q9H0X0, Q9P0W7 | Gene names | SPATA7, HSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 (Spermatogenesis-associated protein HSD3) (HSD-3.1). | |||||
|
SRPK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.008778 (rank : 150) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78362, O75220, O75221, Q6NUL0, Q6V1X2, Q8IYQ3 | Gene names | SRPK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
TFE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.015581 (rank : 131) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
TOP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.014970 (rank : 132) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
ATF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.073688 (rank : 51) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.074503 (rank : 49) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF6A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.086613 (rank : 46) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.077386 (rank : 48) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.073415 (rank : 52) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
CREB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.085959 (rank : 47) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.109425 (rank : 45) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
CREM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.071793 (rank : 54) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |
|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.072403 (rank : 53) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.055118 (rank : 66) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
MAFF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.134262 (rank : 44) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ULX9, Q9Y525 | Gene names | MAFF | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F) (U-Maf). | |||||
|
MAFF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.134944 (rank : 43) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54791 | Gene names | Maff | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.053668 (rank : 70) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 150 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NF2L3_MOUSE
|
||||||
| NC score | 0.971962 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NF2L1_HUMAN
|
||||||
| NC score | 0.890189 (rank : 3) | θ value | 2.47765e-74 (rank : 4) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| NC score | 0.888060 (rank : 4) | θ value | 1.95861e-79 (rank : 3) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
NFE2_HUMAN
|
||||||
| NC score | 0.885162 (rank : 5) | θ value | 1.6774e-38 (rank : 7) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.843105 (rank : 6) | θ value | 5.95217e-44 (rank : 5) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.839513 (rank : 7) | θ value | 2.11701e-41 (rank : 6) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
BACH1_HUMAN
|
||||||
| NC score | 0.389125 (rank : 8) | θ value | 8.95645e-16 (rank : 9) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| NC score | 0.388202 (rank : 9) | θ value | 8.95645e-16 (rank : 10) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
BACH2_MOUSE
|
||||||
| NC score | 0.381698 (rank : 10) | θ value | 6.85773e-16 (rank : 8) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH2_HUMAN
|
||||||
| NC score | 0.378026 (rank : 11) | θ value | 1.52774e-15 (rank : 11) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
JUND_HUMAN
|
||||||
| NC score | 0.348617 (rank : 12) | θ value | 0.00298849 (rank : 18) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.346206 (rank : 13) | θ value | 0.00298849 (rank : 19) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUN_MOUSE
|
||||||
| NC score | 0.344231 (rank : 14) | θ value | 0.00102713 (rank : 15) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUN_HUMAN
|
||||||
| NC score | 0.343621 (rank : 15) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUNB_HUMAN
|
||||||
| NC score | 0.338499 (rank : 16) | θ value | 0.00175202 (rank : 17) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUNB_MOUSE
|
||||||
| NC score | 0.335781 (rank : 17) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
MAFB_MOUSE
|
||||||
| NC score | 0.257415 (rank : 18) | θ value | 0.000158464 (rank : 12) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MAFB_HUMAN
|
||||||
| NC score | 0.256038 (rank : 19) | θ value | 0.00102713 (rank : 16) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
MAFK_HUMAN
|
||||||
| NC score | 0.226192 (rank : 20) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAFK_MOUSE
|
||||||
| NC score | 0.223682 (rank : 21) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAF_MOUSE
|
||||||
| NC score | 0.219538 (rank : 22) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAF_HUMAN
|
||||||
| NC score | 0.217604 (rank : 23) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
XBP1_HUMAN
|
||||||
| NC score | 0.217316 (rank : 24) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
XBP1_MOUSE
|
||||||
| NC score | 0.203274 (rank : 25) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
BATF_MOUSE
|
||||||
| NC score | 0.201524 (rank : 26) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
MAFG_MOUSE
|
||||||
| NC score | 0.199572 (rank : 27) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
MAFG_HUMAN
|
||||||
| NC score | 0.198395 (rank : 28) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15525 | Gene names | MAFG | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G) (hMAF). | |||||
|
ATF3_MOUSE
|
||||||
| NC score | 0.190239 (rank : 29) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
ATF3_HUMAN
|
||||||
| NC score | 0.184922 (rank : 30) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
CREB3_MOUSE
|
||||||
| NC score | 0.184674 (rank : 31) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
BATF_HUMAN
|
||||||
| NC score | 0.183988 (rank : 32) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.172621 (rank : 33) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.170589 (rank : 34) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOS_MOUSE
|
||||||
| NC score | 0.170296 (rank : 35) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.169227 (rank : 36) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOSL2_HUMAN
|
||||||
| NC score | 0.168259 (rank : 37) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL2_MOUSE
|
||||||
| NC score | 0.166948 (rank : 38) | θ value | 0.279714 (rank : 38) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.163336 (rank : 39) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.162399 (rank : 40) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
NRL_MOUSE
|
||||||
| NC score | 0.148865 (rank : 41) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54846 | Gene names | Nrl | |||
|
Domain Architecture |

|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL). | |||||
|
NRL_HUMAN
|
||||||
| NC score | 0.137506 (rank : 42) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
Domain Architecture |

|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
MAFF_MOUSE
|
||||||
| NC score | 0.134944 (rank : 43) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54791 | Gene names | Maff | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F). | |||||
|
MAFF_HUMAN
|
||||||
| NC score | 0.134262 (rank : 44) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ULX9, Q9Y525 | Gene names | MAFF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F) (U-Maf). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.109425 (rank : 45) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.086613 (rank : 46) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
CREB3_HUMAN
|
||||||
| NC score | 0.085959 (rank : 47) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.077386 (rank : 48) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.074503 (rank : 49) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF1_HUMAN
|
||||||
| NC score | 0.074327 (rank : 50) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.073688 (rank : 51) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.073415 (rank : 52) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
CREM_MOUSE
|
||||||
| NC score | 0.072403 (rank : 53) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_HUMAN
|
||||||
| NC score | 0.071793 (rank : 54) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
ATF1_MOUSE
|
||||||
| NC score | 0.071545 (rank : 55) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.070055 (rank : 56) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.069704 (rank : 57) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.067746 (rank : 58) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
S30BP_HUMAN
|
||||||
| NC score | 0.064546 (rank : 59) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHR5, Q8N1R5, Q8TDR8, Q96D27 | Gene names | SAP30BP, HCNGP, HTRG, HTRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAP30-binding protein (Transcriptional regulator protein HCNGP). | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.063730 (rank : 60) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
CREB1_MOUSE
|
||||||
| NC score | 0.060769 (rank : 61) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_HUMAN
|
||||||
| NC score | 0.060749 (rank : 62) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
S30BP_MOUSE
|
||||||
| NC score | 0.060123 (rank : 63) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02614, Q8VDJ5 | Gene names | Sap30bp, Hcngp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAP30-binding protein (Transcriptional regulator protein HCNGP). | |||||
|
SIAL_HUMAN
|
||||||
| NC score | 0.060115 (rank : 64) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |

|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.059346 (rank : 65) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.055118 (rank : 66) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.054773 (rank : 67) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RPO3G_HUMAN
|
||||||
| NC score | 0.054767 (rank : 68) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15318 | Gene names | POLR3G, RPC32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.054657 (rank : 69) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.053668 (rank : 70) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.045750 (rank : 71) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.045165 (rank : 72) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.044153 (rank : 73) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.043856 (rank : 74) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.043683 (rank : 75) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.042747 (rank : 76) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.042344 (rank : 77) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.040901 (rank : 78) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
NCOAT_HUMAN
|
||||||
| NC score | 0.039640 (rank : 79) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
SEMG1_HUMAN
|
||||||
| NC score | 0.039474 (rank : 80) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.038425 (rank : 81) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.038376 (rank : 82) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
LYAR_MOUSE
|
||||||
| NC score | 0.038320 (rank : 83) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
SG16_MOUSE
|
||||||
| NC score | 0.036975 (rank : 84) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.036820 (rank : 85) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.035524 (rank : 86) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
RPO3G_MOUSE
|
||||||
| NC score | 0.035171 (rank : 87) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NXY9, Q8K0W5, Q9CV05 | Gene names | Polr3g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.034371 (rank : 88) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.034176 (rank : 89) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.033585 (rank : 90) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.033572 (rank : 91) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.033510 (rank : 92) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
ARS2_HUMAN
|
||||||
| NC score | 0.033355 (rank : 93) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.033327 (rank : 94) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.032984 (rank : 95) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
TXD13_HUMAN
|
||||||
| NC score | 0.032518 (rank : 96) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.032451 (rank : 97) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.032398 (rank : 98) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.031000 (rank : 99) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
CR025_HUMAN
|
||||||
| NC score | 0.029669 (rank : 100) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96B23, Q5XG78, Q6N058, Q86TB5, Q8TCQ5 | Gene names | C18orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25. | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.029649 (rank : 101) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.028868 (rank : 102) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.027796 (rank : 103) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.027432 (rank : 104) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.025591 (rank : 105) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.025526 (rank : 106) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
IRX2_MOUSE
|
||||||
| NC score | 0.025455 (rank : 107) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81066, O55121, Q9ERN1 | Gene names | Irx2, Irxa2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
PP2CG_HUMAN
|
||||||
| NC score | 0.025099 (rank : 108) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.024741 (rank : 109) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.024240 (rank : 110) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.023989 (rank : 111) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.023747 (rank : 112) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
NDUA7_MOUSE
|
||||||
| NC score | 0.023353 (rank : 113) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z1P6 | Gene names | Ndufa7 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase subunit B14.5a) (Complex I-B14.5a) (CI-B14.5a). | |||||
|
TFE2_HUMAN
|
||||||
| NC score | 0.023110 (rank : 114) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.022894 (rank : 115) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
KI20A_HUMAN
|
||||||
| NC score | 0.021970 (rank : 116) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95235 | Gene names | KIF20A, RAB6KIFL | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin family member 20A (Rabkinesin-6) (Rab6-interacting kinesin- like protein) (GG10_2). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.021671 (rank : 117) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
GET1_HUMAN
|
||||||
| NC score | 0.021480 (rank : 118) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95210 | Gene names | ames=GENX-3414 | |||
|
Domain Architecture |

|
|||||
| Description | Genethonin-1. | |||||
|
UTP20_HUMAN
|
||||||
| NC score | 0.021063 (rank : 119) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
PKP2_HUMAN
|
||||||
| NC score | 0.020781 (rank : 120) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
WDR43_HUMAN
|
||||||
| NC score | 0.020075 (rank : 121) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.019831 (rank : 122) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.018297 (rank : 123) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
SMCE1_HUMAN
|
||||||
| NC score | 0.018136 (rank : 124) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q969G3, O43539 | Gene names | SMARCE1, BAF57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.018034 (rank : 125) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
HDGF_MOUSE
|
||||||
| NC score | 0.016930 (rank : 126) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.016516 (rank : 127) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
RP1_HUMAN
|
||||||
| NC score | 0.016132 (rank : 128) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.016126 (rank : 129) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
TNNT2_HUMAN
|
||||||
| NC score | 0.015956 (rank : 130) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
TFE2_MOUSE
|
||||||
| NC score | 0.015581 (rank : 131) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
TOP1_MOUSE
|
||||||
| NC score | 0.014970 (rank : 132) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
MA1A2_MOUSE
|
||||||
| NC score | 0.014470 (rank : 133) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P39098, Q5SUY6, Q5SUY7, Q60599, Q9CUY9 | Gene names | Man1a2, Man1b | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.013973 (rank : 134) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.013558 (rank : 135) | θ value | 0.21417 (rank : 35) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
R3HD1_HUMAN
|
||||||
| NC score | 0.013275 (rank : 136) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.012189 (rank : 137) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.011923 (rank : 138) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
TXD11_HUMAN
|
||||||
| NC score | 0.011779 (rank : 139) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.011230 (rank : 140) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.011182 (rank : 141) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
SPAT7_HUMAN
|
||||||
| NC score | 0.011010 (rank : 142) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0W8, Q8WX30, Q96HF3, Q9H0X0, Q9P0W7 | Gene names | SPATA7, HSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 (Spermatogenesis-associated protein HSD3) (HSD-3.1). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.010815 (rank : 143) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.010684 (rank : 144) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
KI21B_HUMAN
|
||||||
| NC score | 0.010119 (rank : 145) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
SRPK2_MOUSE
|
||||||
| NC score | 0.009843 (rank : 146) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O54781, Q8VCD9 | Gene names | Srpk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | 0.009669 (rank : 147) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.009563 (rank : 148) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CACB2_MOUSE
|
||||||
| NC score | 0.009437 (rank : 149) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CC27, Q8C5J5, Q9CTQ6 | Gene names | Cacnb2, Cacnlb2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2). | |||||
|
SRPK2_HUMAN
|
||||||
| NC score | 0.008778 (rank : 150) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78362, O75220, O75221, Q6NUL0, Q6V1X2, Q8IYQ3 | Gene names | SRPK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.008290 (rank : 151) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.008257 (rank : 152) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.008214 (rank : 153) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.008191 (rank : 154) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.007855 (rank : 155) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.007623 (rank : 156) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.007509 (rank : 157) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
RNF13_MOUSE
|
||||||
| NC score | 0.006616 (rank : 158) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54965, O54966 | Gene names | Rnf13, Rzf | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 13. | |||||
|
CSTN2_HUMAN
|
||||||
| NC score | 0.006406 (rank : 159) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H4D0, Q9BSS0 | Gene names | CLSTN2, CS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.006267 (rank : 160) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
FOXI1_MOUSE
|
||||||
| NC score | 0.004909 (rank : 161) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q922I5, Q5SRI5, Q9D299 | Gene names | Foxi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I1. | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.004873 (rank : 162) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
PKN2_MOUSE
|
||||||
| NC score | 0.002810 (rank : 163) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BWW9, Q80WS2, Q8BJL7 | Gene names | Pkn2, Prkcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N2 (EC 2.7.11.13) (Protein kinase C- like 2) (Protein-kinase C-related kinase 2). | |||||