Please be patient as the page loads
|
PP2CG_HUMAN
|
||||||
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PP2CG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PP2CG_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.952658 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
PP2CB_MOUSE
|
||||||
| θ value | 1.6813e-30 (rank : 3) | NC score | 0.799481 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P36993 | Gene names | Ppm1b, Pp2c2, Pppm1b | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta) (IA) (Protein phosphatase 1B). | |||||
|
PP2CA_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 4) | NC score | 0.792615 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35813, O75551 | Gene names | PPM1A, PPPM1A | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CB_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 5) | NC score | 0.791354 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75688, Q9HAY8 | Gene names | PPM1B, PP2CB | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta). | |||||
|
PP2CA_MOUSE
|
||||||
| θ value | 7.06379e-29 (rank : 6) | NC score | 0.791001 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49443 | Gene names | Ppm1a, Pppm1a | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CL_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 7) | NC score | 0.774214 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SGD2, Q96NM7 | Gene names | PPM1L, PP2CE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CL_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 8) | NC score | 0.772906 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHN0, Q3UGL2, Q810H0, Q8C021, Q8C1D5, Q9Z0T1 | Gene names | Ppm1l, Kiaa4175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PPM1F_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 9) | NC score | 0.726264 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
PPM1F_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 10) | NC score | 0.730275 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGA0 | Gene names | Ppm1f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Protein phosphatase 1F). | |||||
|
PP2CD_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 11) | NC score | 0.612866 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |
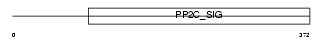
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CD_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 12) | NC score | 0.608940 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZ67 | Gene names | Ppm1d, Wip1 | |||
|
Domain Architecture |
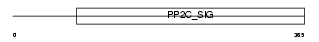
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PDP1_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 13) | NC score | 0.494993 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP2_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 14) | NC score | 0.536245 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
PDP1_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 15) | NC score | 0.487190 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 16) | NC score | 0.199977 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 17) | NC score | 0.199952 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PP2CE_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 18) | NC score | 0.488871 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BU27, Q9CSD6, Q9CU88 | Gene names | Ppm1m, Ppm1e | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
PP2CE_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 19) | NC score | 0.462941 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96MI6, Q8N8J9, Q96DB8 | Gene names | PPM1M, PPM1E | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 20) | NC score | 0.139060 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 21) | NC score | 0.137559 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
PHLPL_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 22) | NC score | 0.158395 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
PHLPL_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 23) | NC score | 0.156095 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 24) | NC score | 0.105474 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TAB1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 25) | NC score | 0.266472 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CF89, Q7TQJ5, Q80V65, Q8R0D1 | Gene names | Map3k7ip1, Tab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 26) | NC score | 0.092879 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TAB1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 27) | NC score | 0.251554 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15750, Q2PP09 | Gene names | MAP3K7IP1, TAB1 | |||
|
Domain Architecture |
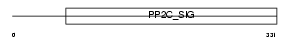
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 28) | NC score | 0.098275 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 29) | NC score | 0.093002 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
F10A1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 30) | NC score | 0.087658 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 31) | NC score | 0.117892 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
CCD47_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 32) | NC score | 0.122837 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96A33, Q96D00, Q96JZ7, Q9H3E4, Q9NRG3 | Gene names | CCDC47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 33) | NC score | 0.084164 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 34) | NC score | 0.085865 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.091917 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
F10A5_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 36) | NC score | 0.081607 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
MIA2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.052013 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZV0 | Gene names | Mia2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.099719 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.015016 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
DEND_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.047764 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
RHPN2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.044201 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWR8, Q9DBN2 | Gene names | Rhpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhophilin-2 (GTP-Rho-binding protein 2). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.061882 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.046061 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.053570 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
LYST_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.039632 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.046139 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
LYST_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.036617 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.042439 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.023345 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
PCIF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.049187 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.033292 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
F10A4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.070316 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
MBP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.058083 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02686, Q15337, Q15338, Q15339, Q15340, Q6AI64, Q6FH37, Q6FI04, Q6PK23 | Gene names | MBP | |||
|
Domain Architecture |
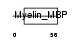
|
|||||
| Description | Myelin basic protein (MBP) (Myelin A1 protein) (Myelin membrane encephalitogenic protein). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.036355 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SMCE1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.037516 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
SYTL5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.021468 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
COBA1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.042517 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
DLG1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.021431 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.002516 (rank : 161) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
NF2L3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.025099 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
PRGR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.009827 (rank : 149) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.034191 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.034200 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.042046 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
DLG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.020651 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.093357 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
KLC4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.026482 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.033572 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RAG2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.034706 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55895, Q8TBL4 | Gene names | RAG2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.031861 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
DEK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.064020 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
TRM6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.039483 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJA5, Q76P92, Q9BQV5, Q9ULR7 | Gene names | TRM6, KIAA1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
AHTF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.022019 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.011164 (rank : 148) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.013881 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.027930 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.044063 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.029123 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.028675 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.022290 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO9A2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.042190 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
CONA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.031390 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
EDAR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.021204 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNE0, Q9UND9 | Gene names | EDAR, DL | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (EDA- A1 receptor) (Ectodermal dysplasia receptor) (Downless homolog). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.043565 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.017068 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.024469 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.009536 (rank : 150) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
POT14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.017646 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.039113 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.026876 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.024449 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
CN004_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.018424 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.081196 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
KLC4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.021112 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |
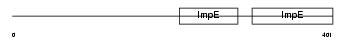
|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.036421 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.056987 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
PLCG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.006184 (rank : 157) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
RBBP5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.022319 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |
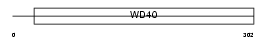
|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.041323 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.019317 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
BNC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.018776 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
BNC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.018997 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.015080 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.024845 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.048461 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.064941 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
RT27_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.024368 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92552 | Gene names | MRPS27, KIAA0264 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 28S ribosomal protein S27 (S27mt) (MRP-S27). | |||||
|
SV2C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.015396 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q496J9, Q496K1, Q9UPU8 | Gene names | SV2C, KIAA1054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle glycoprotein 2C. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.015318 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRI16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.012110 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
TRM6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.031491 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CE96, Q3TJZ8, Q3TME7, Q6ZPW8, Q80Y59, Q8CEU0 | Gene names | Trm6, Kiaa1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
ZN167_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | -0.003704 (rank : 164) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0L1, Q6WL09, Q96FQ2 | Gene names | ZNF167, ZNF448, ZNF64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 167 (Zinc finger protein 64) (Zinc finger protein 448) (ZFP). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.019770 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
AT1A3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.005662 (rank : 159) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.005667 (rank : 158) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
BPIL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.018263 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C186 | Gene names | Bpil2 | |||
|
Domain Architecture |
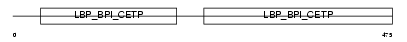
|
|||||
| Description | Bactericidal/permeability-increasing protein-like 2 precursor. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.082698 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.043473 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GAK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.000252 (rank : 162) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
HDAC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.006638 (rank : 156) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15379, O43268, Q9UEI5, Q9UEV0 | Gene names | HDAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 3 (HD3) (RPD3-2) (SMAP45). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | -0.001045 (rank : 163) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.018706 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
SAFB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.014347 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.007171 (rank : 154) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.054041 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
TM16H_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.020558 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
ARHGA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.006916 (rank : 155) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
BPIL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.015680 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFQ6 | Gene names | BPIL2 | |||
|
Domain Architecture |
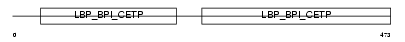
|
|||||
| Description | Bactericidal/permeability-increasing protein-like 2 precursor. | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.025211 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CT175_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.015655 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MK2, Q5QPB6, Q9NQQ2 | Gene names | C20orf175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf175. | |||||
|
DLG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.014689 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.014713 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.008039 (rank : 153) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
GTR9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.005211 (rank : 160) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRM0 | Gene names | SLC2A9, GLUT9 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 9 (Glucose transporter type 9). | |||||
|
HOME1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.008067 (rank : 152) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
MY116_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.028822 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17564 | Gene names | Myd116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid differentiation primary response protein MyD116. | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.018979 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
RAG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.025664 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.012665 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
TSH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.008396 (rank : 151) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.051898 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
CENPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.052298 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.060152 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.059971 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.054039 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.069608 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.072713 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.065734 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.094324 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.059928 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.057971 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.053871 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.125876 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.125574 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.089822 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.098930 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.053181 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.053893 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.057511 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.059555 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.052635 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.051011 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.050130 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.061625 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
PP2CG_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PP2CG_MOUSE
|
||||||
| NC score | 0.952658 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
PP2CB_MOUSE
|
||||||
| NC score | 0.799481 (rank : 3) | θ value | 1.6813e-30 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P36993 | Gene names | Ppm1b, Pp2c2, Pppm1b | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta) (IA) (Protein phosphatase 1B). | |||||
|
PP2CA_HUMAN
|
||||||
| NC score | 0.792615 (rank : 4) | θ value | 2.86786e-30 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35813, O75551 | Gene names | PPM1A, PPPM1A | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CB_HUMAN
|
||||||
| NC score | 0.791354 (rank : 5) | θ value | 1.85889e-29 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75688, Q9HAY8 | Gene names | PPM1B, PP2CB | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta). | |||||
|
PP2CA_MOUSE
|
||||||
| NC score | 0.791001 (rank : 6) | θ value | 7.06379e-29 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49443 | Gene names | Ppm1a, Pppm1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CL_HUMAN
|
||||||
| NC score | 0.774214 (rank : 7) | θ value | 4.15078e-21 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SGD2, Q96NM7 | Gene names | PPM1L, PP2CE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CL_MOUSE
|
||||||
| NC score | 0.772906 (rank : 8) | θ value | 7.0802e-21 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHN0, Q3UGL2, Q810H0, Q8C021, Q8C1D5, Q9Z0T1 | Gene names | Ppm1l, Kiaa4175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PPM1F_MOUSE
|
||||||
| NC score | 0.730275 (rank : 9) | θ value | 2.7842e-17 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGA0 | Gene names | Ppm1f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Protein phosphatase 1F). | |||||
|
PPM1F_HUMAN
|
||||||
| NC score | 0.726264 (rank : 10) | θ value | 1.92812e-18 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
PP2CD_HUMAN
|
||||||
| NC score | 0.612866 (rank : 11) | θ value | 7.84624e-12 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CD_MOUSE
|
||||||
| NC score | 0.608940 (rank : 12) | θ value | 2.28291e-11 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZ67 | Gene names | Ppm1d, Wip1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PDP2_HUMAN
|
||||||
| NC score | 0.536245 (rank : 13) | θ value | 2.13673e-09 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
PDP1_MOUSE
|
||||||
| NC score | 0.494993 (rank : 14) | θ value | 2.13673e-09 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PP2CE_MOUSE
|
||||||
| NC score | 0.488871 (rank : 15) | θ value | 6.43352e-06 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BU27, Q9CSD6, Q9CU88 | Gene names | Ppm1m, Ppm1e | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
PDP1_HUMAN
|
||||||
| NC score | 0.487190 (rank : 16) | θ value | 8.11959e-09 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PP2CE_HUMAN
|
||||||
| NC score | 0.462941 (rank : 17) | θ value | 7.1131e-05 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96MI6, Q8N8J9, Q96DB8 | Gene names | PPM1M, PPM1E | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
TAB1_MOUSE
|
||||||
| NC score | 0.266472 (rank : 18) | θ value | 0.00390308 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CF89, Q7TQJ5, Q80V65, Q8R0D1 | Gene names | Map3k7ip1, Tab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
TAB1_HUMAN
|
||||||
| NC score | 0.251554 (rank : 19) | θ value | 0.00509761 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15750, Q2PP09 | Gene names | MAP3K7IP1, TAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.199977 (rank : 20) | θ value | 1.17247e-07 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.199952 (rank : 21) | θ value | 7.59969e-07 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PHLPL_HUMAN
|
||||||
| NC score | 0.158395 (rank : 22) | θ value | 0.00298849 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
PHLPL_MOUSE
|
||||||
| NC score | 0.156095 (rank : 23) | θ value | 0.00298849 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.139060 (rank : 24) | θ value | 0.000786445 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.137559 (rank : 25) | θ value | 0.00102713 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.125876 (rank : 26) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.125574 (rank : 27) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
CCD47_HUMAN
|
||||||
| NC score | 0.122837 (rank : 28) | θ value | 0.0193708 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96A33, Q96D00, Q96JZ7, Q9H3E4, Q9NRG3 | Gene names | CCDC47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor. | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.117892 (rank : 29) | θ value | 0.0193708 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.105474 (rank : 30) | θ value | 0.00390308 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.099719 (rank : 31) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.098930 (rank : 32) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.098275 (rank : 33) | θ value | 0.00869519 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.094324 (rank : 34) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.093357 (rank : 35) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.093002 (rank : 36) | θ value | 0.0113563 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.092879 (rank : 37) | θ value | 0.00509761 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.091917 (rank : 38) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.089822 (rank : 39) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.087658 (rank : 40) | θ value | 0.0148317 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.085865 (rank : 41) | θ value | 0.0563607 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.084164 (rank : 42) | θ value | 0.0330416 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.082698 (rank : 43) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.081607 (rank : 44) | θ value | 0.0736092 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.081196 (rank : 45) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.072713 (rank : 46) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
F10A4_HUMAN
|
||||||
| NC score | 0.070316 (rank : 47) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IZP2 | Gene names | FAM10A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A4. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.069608 (rank : 48) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.065734 (rank : 49) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.064941 (rank : 50) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.064020 (rank : 51) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.061882 (rank : 52) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.061625 (rank : 53) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.060152 (rank : 54) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.059971 (rank : 55) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.059928 (rank : 56) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.059555 (rank : 57) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
MBP_HUMAN
|
||||||
| NC score | 0.058083 (rank : 58) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02686, Q15337, Q15338, Q15339, Q15340, Q6AI64, Q6FH37, Q6FI04, Q6PK23 | Gene names | MBP | |||
|
Domain Architecture |

|
|||||
| Description | Myelin basic protein (MBP) (Myelin A1 protein) (Myelin membrane encephalitogenic protein). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.057971 (rank : 59) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.057511 (rank : 60) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.056987 (rank : 61) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.054041 (rank : 62) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.054039 (rank : 63) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.053893 (rank : 64) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.053871 (rank : 65) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.053570 (rank : 66) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.053181 (rank : 67) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.052635 (rank : 68) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
CENPB_HUMAN
|
||||||
| NC score | 0.052298 (rank : 69) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
MIA2_MOUSE
|
||||||
| NC score | 0.052013 (rank : 70) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZV0 | Gene names | Mia2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma inhibitory activity protein 2 precursor. | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.051898 (rank : 71) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.051011 (rank : 72) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.050130 (rank : 73) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
PCIF1_HUMAN
|
||||||
| NC score | 0.049187 (rank : 74) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.048461 (rank : 75) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
DEND_HUMAN
|
||||||
| NC score | 0.047764 (rank : 76) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94850 | Gene names | DDN, KIAA0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.046139 (rank : 77) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.046061 (rank : 78) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
RHPN2_MOUSE
|
||||||
| NC score | 0.044201 (rank : 79) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWR8, Q9DBN2 | Gene names | Rhpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhophilin-2 (GTP-Rho-binding protein 2). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.044063 (rank : 80) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.043565 (rank : 81) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.043473 (rank : 82) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.042517 (rank : 83) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.042439 (rank : 84) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
CO9A2_MOUSE
|
||||||
| NC score | 0.042190 (rank : 85) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.042046 (rank : 86) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.041323 (rank : 87) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.039632 (rank : 88) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
TRM6_HUMAN
|
||||||
| NC score | 0.039483 (rank : 89) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJA5, Q76P92, Q9BQV5, Q9ULR7 | Gene names | TRM6, KIAA1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.039113 (rank : 90) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SMCE1_MOUSE
|
||||||
| NC score | 0.037516 (rank : 91) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O54941, Q8BPD9 | Gene names | Smarce1, Baf57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator chromatin subfamily E member 1 (BRG1-associated factor 57). | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.036617 (rank : 92) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.036421 (rank : 93) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.036355 (rank : 94) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
RAG2_HUMAN
|
||||||
| NC score | 0.034706 (rank : 95) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55895, Q8TBL4 | Gene names | RAG2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.034200 (rank : 96) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.034191 (rank : 97) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.033572 (rank : 98) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.033292 (rank : 99) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.031861 (rank : 100) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
TRM6_MOUSE
|
||||||
| NC score | 0.031491 (rank : 101) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CE96, Q3TJZ8, Q3TME7, Q6ZPW8, Q80Y59, Q8CEU0 | Gene names | Trm6, Kiaa1153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA (adenine-N(1)-)-methyltransferase non-catalytic subunit TRM6 (tRNA(m1A58)-methyltransferase subunit TRM6) (tRNA(m1A58)MTase subunit TRM6). | |||||
|
CONA1_HUMAN
|
||||||
| NC score | 0.031390 (rank : 102) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.029123 (rank : 103) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
MY116_MOUSE
|
||||||
| NC score | 0.028822 (rank : 104) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17564 | Gene names | Myd116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid differentiation primary response protein MyD116. | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.028675 (rank : 105) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.027930 (rank : 106) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.026876 (rank : 107) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.026482 (rank : 108) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
RAG2_MOUSE
|
||||||
| NC score | 0.025664 (rank : 109) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21784 | Gene names | Rag2, Rag-2 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 2 (RAG-2). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.025211 (rank : 110) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.025099 (rank : 111) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.024845 (rank : 112) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.024469 (rank : 113) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.024449 (rank : 114) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
RT27_HUMAN
|
||||||
| NC score | 0.024368 (rank : 115) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92552 | Gene names | MRPS27, KIAA0264 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial 28S ribosomal protein S27 (S27mt) (MRP-S27). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.023345 (rank : 116) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
RBBP5_HUMAN
|
||||||
| NC score | 0.022319 (rank : 117) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15291 | Gene names | RBBP5, RBQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.022290 (rank : 118) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
AHTF1_MOUSE
|
||||||
| NC score | 0.022019 (rank : 119) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
SYTL5_HUMAN
|
||||||
| NC score | 0.021468 (rank : 120) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
DLG1_MOUSE
|
||||||
| NC score | 0.021431 (rank : 121) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
EDAR_HUMAN
|
||||||
| NC score | 0.021204 (rank : 122) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UNE0, Q9UND9 | Gene names | EDAR, DL | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (EDA- A1 receptor) (Ectodermal dysplasia receptor) (Downless homolog). | |||||
|
KLC4_MOUSE
|
||||||
| NC score | 0.021112 (rank : 123) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBS5, Q3TCC5 | Gene names | Klc4, Knsl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
DLG1_HUMAN
|
||||||
| NC score | 0.020651 (rank : 124) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
TM16H_HUMAN
|
||||||
| NC score | 0.020558 (rank : 125) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HCE9 | Gene names | TMEM16H, KIAA1623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 16H. | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.019770 (rank : 126) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.019317 (rank : 127) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
BNC2_MOUSE
|
||||||
| NC score | 0.018997 (rank : 128) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.018979 (rank : 129) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
BNC2_HUMAN
|
||||||
| NC score | 0.018776 (rank : 130) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.018706 (rank : 131) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.018424 (rank : 132) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
BPIL2_MOUSE
|
||||||
| NC score | 0.018263 (rank : 133) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C186 | Gene names | Bpil2 | |||
|
Domain Architecture |

|
|||||
| Description | Bactericidal/permeability-increasing protein-like 2 precursor. | |||||
|
POT14_HUMAN
|
||||||
| NC score | 0.017646 (rank : 134) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.017068 (rank : 135) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
BPIL2_HUMAN
|
||||||
| NC score | 0.015680 (rank : 136) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFQ6 | Gene names | BPIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Bactericidal/permeability-increasing protein-like 2 precursor. | |||||
|
CT175_HUMAN
|
||||||
| NC score | 0.015655 (rank : 137) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MK2, Q5QPB6, Q9NQQ2 | Gene names | C20orf175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf175. | |||||
|
SV2C_HUMAN
|
||||||
| NC score | 0.015396 (rank : 138) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q496J9, Q496K1, Q9UPU8 | Gene names | SV2C, KIAA1054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle glycoprotein 2C. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.015318 (rank : 139) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.015080 (rank : 140) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.015016 (rank : 141) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
DLG4_MOUSE
|
||||||
| NC score | 0.014713 (rank : 142) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG4_HUMAN
|
||||||
| NC score | 0.014689 (rank : 143) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
SAFB2_HUMAN
|
||||||
| NC score | 0.014347 (rank : 144) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.013881 (rank : 145) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.012665 (rank : 146) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
TRI16_HUMAN
|
||||||
| NC score | 0.012110 (rank : 147) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.011164 (rank : 148) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
PRGR_HUMAN
|
||||||
| NC score | 0.009827 (rank : 149) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.009536 (rank : 150) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
TSH2_HUMAN
|
||||||
| NC score | 0.008396 (rank : 151) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
HOME1_MOUSE
|
||||||
| NC score | 0.008067 (rank : 152) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.008039 (rank : 153) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.007171 (rank : 154) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
ARHGA_HUMAN
|
||||||
| NC score | 0.006916 (rank : 155) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
HDAC3_HUMAN
|
||||||
| NC score | 0.006638 (rank : 156) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15379, O43268, Q9UEI5, Q9UEV0 | Gene names | HDAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 3 (HD3) (RPD3-2) (SMAP45). | |||||
|
PLCG1_MOUSE
|
||||||
| NC score | 0.006184 (rank : 157) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
AT1A3_MOUSE
|
||||||
| NC score | 0.005667 (rank : 158) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PIC6 | Gene names | Atp1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
AT1A3_HUMAN
|
||||||
| NC score | 0.005662 (rank : 159) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13637, Q16732, Q16735, Q969K5 | Gene names | ATP1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium-transporting ATPase alpha-3 chain (EC 3.6.3.9) (Sodium pump 3) (Na+/K+ ATPase 3) (Alpha(III)). | |||||
|
GTR9_HUMAN
|
||||||
| NC score | 0.005211 (rank : 160) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRM0 | Gene names | SLC2A9, GLUT9 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 9 (Glucose transporter type 9). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.002516 (rank : 161) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.000252 (rank : 162) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | -0.001045 (rank : 163) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ZN167_HUMAN
|
||||||
| NC score | -0.003704 (rank : 164) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0L1, Q6WL09, Q96FQ2 | Gene names | ZNF167, ZNF448, ZNF64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 167 (Zinc finger protein 64) (Zinc finger protein 448) (ZFP). | |||||