Please be patient as the page loads
|
PP2CD_HUMAN
|
||||||
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PP2CD_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CD_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995479 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZ67 | Gene names | Ppm1d, Wip1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CA_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 3) | NC score | 0.674324 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35813, O75551 | Gene names | PPM1A, PPPM1A | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CB_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 4) | NC score | 0.673512 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P36993 | Gene names | Ppm1b, Pp2c2, Pppm1b | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta) (IA) (Protein phosphatase 1B). | |||||
|
PP2CB_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 5) | NC score | 0.673068 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75688, Q9HAY8 | Gene names | PPM1B, PP2CB | |||
|
Domain Architecture |
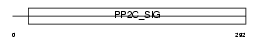
|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta). | |||||
|
PP2CA_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 6) | NC score | 0.671543 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49443 | Gene names | Ppm1a, Pppm1a | |||
|
Domain Architecture |
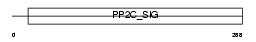
|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PPM1F_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 7) | NC score | 0.661968 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
PPM1F_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 8) | NC score | 0.650519 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CGA0 | Gene names | Ppm1f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Protein phosphatase 1F). | |||||
|
PP2CL_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 9) | NC score | 0.636447 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SGD2, Q96NM7 | Gene names | PPM1L, PP2CE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CL_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 10) | NC score | 0.634807 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHN0, Q3UGL2, Q810H0, Q8C021, Q8C1D5, Q9Z0T1 | Gene names | Ppm1l, Kiaa4175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CG_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 11) | NC score | 0.612866 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PP2CG_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 12) | NC score | 0.596210 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
PDP1_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 13) | NC score | 0.468569 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP1_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 14) | NC score | 0.472287 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP2_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 15) | NC score | 0.490956 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
PP2CE_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 16) | NC score | 0.386317 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BU27, Q9CSD6, Q9CU88 | Gene names | Ppm1m, Ppm1e | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.022464 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.021009 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
ANR25_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.011634 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
COX10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.027837 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.022097 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
POPD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.021799 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ES82 | Gene names | Popdc2, Pop2 | |||
|
Domain Architecture |

|
|||||
| Description | Popeye domain-containing protein 2 (Popeye protein 2). | |||||
|
SACS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.021529 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.018539 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
LPHN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.007987 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95490, O94882, Q5VX76, Q9UKY5, Q9UKY6 | Gene names | LPHN2, KIAA0786, LEC1, LPHH1 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-2 precursor (Calcium-independent alpha-latrotoxin receptor 2) (Latrophilin homolog 1) (Lectomedin-1). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.013747 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.013171 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SSH2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.012130 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.000118 (rank : 39) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.014909 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.008372 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.011992 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.005341 (rank : 37) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.005302 (rank : 38) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.068026 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.069608 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PP2CE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.282452 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96MI6, Q8N8J9, Q96DB8 | Gene names | PPM1M, PPM1E | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
TAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.109216 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15750, Q2PP09 | Gene names | MAP3K7IP1, TAB1 | |||
|
Domain Architecture |
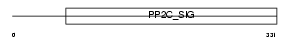
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
TAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.122095 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CF89, Q7TQJ5, Q80V65, Q8R0D1 | Gene names | Map3k7ip1, Tab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
PP2CD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CD_MOUSE
|
||||||
| NC score | 0.995479 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZ67 | Gene names | Ppm1d, Wip1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CA_HUMAN
|
||||||
| NC score | 0.674324 (rank : 3) | θ value | 4.74913e-17 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35813, O75551 | Gene names | PPM1A, PPPM1A | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CB_MOUSE
|
||||||
| NC score | 0.673512 (rank : 4) | θ value | 6.20254e-17 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P36993 | Gene names | Ppm1b, Pp2c2, Pppm1b | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta) (IA) (Protein phosphatase 1B). | |||||
|
PP2CB_HUMAN
|
||||||
| NC score | 0.673068 (rank : 5) | θ value | 8.10077e-17 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75688, Q9HAY8 | Gene names | PPM1B, PP2CB | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta). | |||||
|
PP2CA_MOUSE
|
||||||
| NC score | 0.671543 (rank : 6) | θ value | 1.058e-16 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49443 | Gene names | Ppm1a, Pppm1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PPM1F_HUMAN
|
||||||
| NC score | 0.661968 (rank : 7) | θ value | 2.20605e-14 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
PPM1F_MOUSE
|
||||||
| NC score | 0.650519 (rank : 8) | θ value | 1.42992e-13 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CGA0 | Gene names | Ppm1f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Protein phosphatase 1F). | |||||
|
PP2CL_HUMAN
|
||||||
| NC score | 0.636447 (rank : 9) | θ value | 4.16044e-13 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SGD2, Q96NM7 | Gene names | PPM1L, PP2CE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CL_MOUSE
|
||||||
| NC score | 0.634807 (rank : 10) | θ value | 5.43371e-13 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHN0, Q3UGL2, Q810H0, Q8C021, Q8C1D5, Q9Z0T1 | Gene names | Ppm1l, Kiaa4175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CG_HUMAN
|
||||||
| NC score | 0.612866 (rank : 11) | θ value | 7.84624e-12 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PP2CG_MOUSE
|
||||||
| NC score | 0.596210 (rank : 12) | θ value | 7.84624e-12 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
PDP2_HUMAN
|
||||||
| NC score | 0.490956 (rank : 13) | θ value | 3.41135e-07 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
PDP1_MOUSE
|
||||||
| NC score | 0.472287 (rank : 14) | θ value | 6.21693e-09 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP1_HUMAN
|
||||||
| NC score | 0.468569 (rank : 15) | θ value | 6.21693e-09 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PP2CE_MOUSE
|
||||||
| NC score | 0.386317 (rank : 16) | θ value | 0.00175202 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BU27, Q9CSD6, Q9CU88 | Gene names | Ppm1m, Ppm1e | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
PP2CE_HUMAN
|
||||||
| NC score | 0.282452 (rank : 17) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96MI6, Q8N8J9, Q96DB8 | Gene names | PPM1M, PPM1E | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
TAB1_MOUSE
|
||||||
| NC score | 0.122095 (rank : 18) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CF89, Q7TQJ5, Q80V65, Q8R0D1 | Gene names | Map3k7ip1, Tab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
TAB1_HUMAN
|
||||||
| NC score | 0.109216 (rank : 19) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15750, Q2PP09 | Gene names | MAP3K7IP1, TAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.069608 (rank : 20) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.068026 (rank : 21) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
COX10_MOUSE
|
||||||
| NC score | 0.027837 (rank : 22) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.022464 (rank : 23) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.022097 (rank : 24) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
POPD2_MOUSE
|
||||||
| NC score | 0.021799 (rank : 25) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ES82 | Gene names | Popdc2, Pop2 | |||
|
Domain Architecture |

|
|||||
| Description | Popeye domain-containing protein 2 (Popeye protein 2). | |||||
|
SACS_MOUSE
|
||||||
| NC score | 0.021529 (rank : 26) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLC8 | Gene names | Sacs | |||
|
Domain Architecture |

|
|||||
| Description | Sacsin. | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.021009 (rank : 27) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.018539 (rank : 28) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.014909 (rank : 29) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.013747 (rank : 30) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.013171 (rank : 31) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SSH2_MOUSE
|
||||||
| NC score | 0.012130 (rank : 32) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.011992 (rank : 33) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
ANR25_MOUSE
|
||||||
| NC score | 0.011634 (rank : 34) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.008372 (rank : 35) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
LPHN2_HUMAN
|
||||||
| NC score | 0.007987 (rank : 36) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95490, O94882, Q5VX76, Q9UKY5, Q9UKY6 | Gene names | LPHN2, KIAA0786, LEC1, LPHH1 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-2 precursor (Calcium-independent alpha-latrotoxin receptor 2) (Latrophilin homolog 1) (Lectomedin-1). | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.005341 (rank : 37) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.005302 (rank : 38) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.000118 (rank : 39) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||