Please be patient as the page loads
|
PDP1_MOUSE
|
||||||
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PDP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999656 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP2_HUMAN
|
||||||
| θ value | 7.30591e-143 (rank : 3) | NC score | 0.968718 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
PP2CG_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 4) | NC score | 0.494993 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PP2CG_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 5) | NC score | 0.480991 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
PP2CD_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 6) | NC score | 0.472287 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |
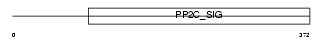
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CL_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 7) | NC score | 0.499646 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SGD2, Q96NM7 | Gene names | PPM1L, PP2CE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CL_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 8) | NC score | 0.497903 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHN0, Q3UGL2, Q810H0, Q8C021, Q8C1D5, Q9Z0T1 | Gene names | Ppm1l, Kiaa4175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CD_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 9) | NC score | 0.458871 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZ67 | Gene names | Ppm1d, Wip1 | |||
|
Domain Architecture |
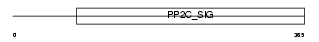
|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CE_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 10) | NC score | 0.454035 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BU27, Q9CSD6, Q9CU88 | Gene names | Ppm1m, Ppm1e | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
PP2CB_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 11) | NC score | 0.428549 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75688, Q9HAY8 | Gene names | PPM1B, PP2CB | |||
|
Domain Architecture |
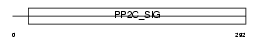
|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta). | |||||
|
PP2CB_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 12) | NC score | 0.427946 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P36993 | Gene names | Ppm1b, Pp2c2, Pppm1b | |||
|
Domain Architecture |
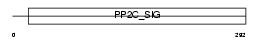
|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta) (IA) (Protein phosphatase 1B). | |||||
|
PP2CA_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 13) | NC score | 0.405645 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35813, O75551 | Gene names | PPM1A, PPPM1A | |||
|
Domain Architecture |
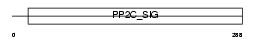
|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CA_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 14) | NC score | 0.404921 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49443 | Gene names | Ppm1a, Pppm1a | |||
|
Domain Architecture |
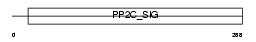
|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PPM1F_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 15) | NC score | 0.407519 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
PP2CE_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 16) | NC score | 0.379963 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96MI6, Q8N8J9, Q96DB8 | Gene names | PPM1M, PPM1E | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
TAB1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 17) | NC score | 0.234876 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15750, Q2PP09 | Gene names | MAP3K7IP1, TAB1 | |||
|
Domain Architecture |
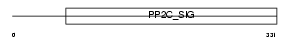
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
TAB1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 18) | NC score | 0.272099 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CF89, Q7TQJ5, Q80V65, Q8R0D1 | Gene names | Map3k7ip1, Tab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
PPM1F_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.384366 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGA0 | Gene names | Ppm1f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Protein phosphatase 1F). | |||||
|
PHLPL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.087274 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
PHLPL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.083649 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.100736 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.098842 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
CJ119_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.058656 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3C0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119 homolog. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.006472 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
KR151_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.025052 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI76 | Gene names | KRTAP15-1, KAP15.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 15-1. | |||||
|
ODFP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.036363 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |
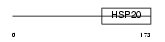
|
|||||
| Description | Outer dense fiber protein. | |||||
|
SYIC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.025430 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
HS70A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.020982 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.020982 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.020897 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
CJ119_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.043184 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BTE3, Q6IA56, Q9BVT9, Q9H916 | Gene names | C10orf119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119. | |||||
|
HSP71_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.019893 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
FANCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.020574 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15360, O75266, Q6PL10, Q92497, Q96H18, Q9UEA5, Q9UEL8, Q9UEL9, Q9UPK3, Q9Y6M2 | Gene names | FANCA, FAA | |||
|
Domain Architecture |

|
|||||
| Description | Fanconi anemia group A protein (Protein FACA). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.009084 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
ADCK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.019063 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0L4, Q3UKJ2 | Gene names | Adck1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized aarF domain-containing protein kinase 1 precursor (EC 2.7.-.-). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.003902 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
PDP1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP1_HUMAN
|
||||||
| NC score | 0.999656 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP2_HUMAN
|
||||||
| NC score | 0.968718 (rank : 3) | θ value | 7.30591e-143 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
PP2CL_HUMAN
|
||||||
| NC score | 0.499646 (rank : 4) | θ value | 4.0297e-08 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SGD2, Q96NM7 | Gene names | PPM1L, PP2CE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CL_MOUSE
|
||||||
| NC score | 0.497903 (rank : 5) | θ value | 5.26297e-08 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHN0, Q3UGL2, Q810H0, Q8C021, Q8C1D5, Q9Z0T1 | Gene names | Ppm1l, Kiaa4175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 2C isoform epsilon (EC 3.1.3.16) (PP2C-epsilon) (Protein phosphatase 1L) (Protein phosphatase 1-like). | |||||
|
PP2CG_HUMAN
|
||||||
| NC score | 0.494993 (rank : 6) | θ value | 2.13673e-09 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15355 | Gene names | PPM1G, PPM1C | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C). | |||||
|
PP2CG_MOUSE
|
||||||
| NC score | 0.480991 (rank : 7) | θ value | 2.13673e-09 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
PP2CD_HUMAN
|
||||||
| NC score | 0.472287 (rank : 8) | θ value | 6.21693e-09 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15297 | Gene names | PPM1D, WIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CD_MOUSE
|
||||||
| NC score | 0.458871 (rank : 9) | θ value | 8.97725e-08 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZ67 | Gene names | Ppm1d, Wip1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform delta (EC 3.1.3.16) (PP2C-delta) (p53- induced protein phosphatase 1) (Protein phosphatase magnesium- dependent 1 delta). | |||||
|
PP2CE_MOUSE
|
||||||
| NC score | 0.454035 (rank : 10) | θ value | 5.81887e-07 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BU27, Q9CSD6, Q9CU88 | Gene names | Ppm1m, Ppm1e | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
PP2CB_HUMAN
|
||||||
| NC score | 0.428549 (rank : 11) | θ value | 3.77169e-06 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75688, Q9HAY8 | Gene names | PPM1B, PP2CB | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta). | |||||
|
PP2CB_MOUSE
|
||||||
| NC score | 0.427946 (rank : 12) | θ value | 2.44474e-05 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P36993 | Gene names | Ppm1b, Pp2c2, Pppm1b | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform beta (EC 3.1.3.16) (PP2C-beta) (IA) (Protein phosphatase 1B). | |||||
|
PPM1F_HUMAN
|
||||||
| NC score | 0.407519 (rank : 13) | θ value | 0.00035302 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
PP2CA_HUMAN
|
||||||
| NC score | 0.405645 (rank : 14) | θ value | 0.00020696 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35813, O75551 | Gene names | PPM1A, PPPM1A | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PP2CA_MOUSE
|
||||||
| NC score | 0.404921 (rank : 15) | θ value | 0.00020696 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49443 | Gene names | Ppm1a, Pppm1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform alpha (EC 3.1.3.16) (PP2C-alpha) (IA) (Protein phosphatase 1A). | |||||
|
PPM1F_MOUSE
|
||||||
| NC score | 0.384366 (rank : 16) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CGA0 | Gene names | Ppm1f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Protein phosphatase 1F). | |||||
|
PP2CE_HUMAN
|
||||||
| NC score | 0.379963 (rank : 17) | θ value | 0.000461057 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96MI6, Q8N8J9, Q96DB8 | Gene names | PPM1M, PPM1E | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform eta (EC 3.1.3.16) (PP2C-eta) (PP2CE) (Protein phosphatase 1M). | |||||
|
TAB1_MOUSE
|
||||||
| NC score | 0.272099 (rank : 18) | θ value | 0.00228821 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CF89, Q7TQJ5, Q80V65, Q8R0D1 | Gene names | Map3k7ip1, Tab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
TAB1_HUMAN
|
||||||
| NC score | 0.234876 (rank : 19) | θ value | 0.00228821 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15750, Q2PP09 | Gene names | MAP3K7IP1, TAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 1 (TGF-beta-activated kinase 1-binding protein 1) (TAK1-binding protein 1). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.100736 (rank : 20) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.098842 (rank : 21) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PHLPL_HUMAN
|
||||||
| NC score | 0.087274 (rank : 22) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
PHLPL_MOUSE
|
||||||
| NC score | 0.083649 (rank : 23) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXA7, Q8BX96 | Gene names | Phlppl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
CJ119_MOUSE
|
||||||
| NC score | 0.058656 (rank : 24) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3C0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119 homolog. | |||||
|
CJ119_HUMAN
|
||||||
| NC score | 0.043184 (rank : 25) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BTE3, Q6IA56, Q9BVT9, Q9H916 | Gene names | C10orf119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119. | |||||
|
ODFP_MOUSE
|
||||||
| NC score | 0.036363 (rank : 26) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
SYIC_MOUSE
|
||||||
| NC score | 0.025430 (rank : 27) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
KR151_HUMAN
|
||||||
| NC score | 0.025052 (rank : 28) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3LI76 | Gene names | KRTAP15-1, KAP15.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 15-1. | |||||
|
HS70B_MOUSE
|
||||||
| NC score | 0.020982 (rank : 29) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70A_MOUSE
|
||||||
| NC score | 0.020982 (rank : 30) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70L_HUMAN
|
||||||
| NC score | 0.020897 (rank : 31) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
FANCA_HUMAN
|
||||||
| NC score | 0.020574 (rank : 32) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15360, O75266, Q6PL10, Q92497, Q96H18, Q9UEA5, Q9UEL8, Q9UEL9, Q9UPK3, Q9Y6M2 | Gene names | FANCA, FAA | |||
|
Domain Architecture |

|
|||||
| Description | Fanconi anemia group A protein (Protein FACA). | |||||
|
HSP71_HUMAN
|
||||||
| NC score | 0.019893 (rank : 33) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
ADCK1_MOUSE
|
||||||
| NC score | 0.019063 (rank : 34) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0L4, Q3UKJ2 | Gene names | Adck1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized aarF domain-containing protein kinase 1 precursor (EC 2.7.-.-). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.009084 (rank : 35) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.006472 (rank : 36) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.003902 (rank : 37) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||