Please be patient as the page loads
|
HS70B_MOUSE
|
||||||
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GRP78_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.983557 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.983454 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
HS70A_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999929 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70L_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.997341 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HS70L_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.997594 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HSP71_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.998510 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HSP72_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.996792 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP72_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.996529 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP76_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.994867 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP7C_HUMAN
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.993321 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.992668 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 1.92261e-143 (rank : 13) | NC score | 0.981857 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 2.77624e-142 (rank : 14) | NC score | 0.982710 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
HS74L_MOUSE
|
||||||
| θ value | 2.92676e-75 (rank : 15) | NC score | 0.902734 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 8.51556e-75 (rank : 16) | NC score | 0.891054 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
HSP74_HUMAN
|
||||||
| θ value | 1.45253e-74 (rank : 17) | NC score | 0.902604 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
HS74L_HUMAN
|
||||||
| θ value | 1.22964e-73 (rank : 18) | NC score | 0.901311 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
STCH_HUMAN
|
||||||
| θ value | 2.09745e-73 (rank : 19) | NC score | 0.963460 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_MOUSE
|
||||||
| θ value | 1.35953e-72 (rank : 20) | NC score | 0.963131 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS105_HUMAN
|
||||||
| θ value | 1.50314e-71 (rank : 21) | NC score | 0.898144 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HS105_MOUSE
|
||||||
| θ value | 1.96316e-71 (rank : 22) | NC score | 0.905607 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 9.16162e-53 (rank : 23) | NC score | 0.874423 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 24) | NC score | 0.253812 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 25) | NC score | 0.144997 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
ANR45_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 26) | NC score | 0.194545 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 27) | NC score | 0.045496 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
E2AK2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.007519 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |
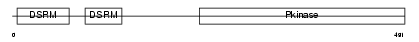
|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
HS12B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 29) | NC score | 0.090995 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 30) | NC score | 0.041816 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.041469 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.027506 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.040984 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.039534 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PCIF1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.065451 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
PCIF1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.057120 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59114 | Gene names | Pcif1 | |||
|
Domain Architecture |
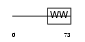
|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.038383 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.027816 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
COVA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.062069 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |
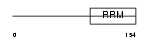
|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.057481 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
K2C3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.018732 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |
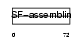
|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
COVA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.050289 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.027888 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.018510 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.029947 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.046249 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
DZIP3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.018808 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.033359 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.029949 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.025871 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.041431 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.031004 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.021836 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.022104 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.026296 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
PDP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.021063 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.020982 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.016464 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.023035 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.025338 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
FLNA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.023324 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
K2C7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.011916 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08729, Q92676, Q9BUD8, Q9Y3R7 | Gene names | KRT7, SCL | |||
|
Domain Architecture |
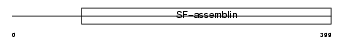
|
|||||
| Description | Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.033275 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.029023 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
UBXD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.034722 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
UBXD2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.032176 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.056045 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.017778 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
K2C1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.011936 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
APOL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.010027 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQE5, O95915, Q5TH96, Q969T6, Q9BT28, Q9UGT1, Q9UH10 | Gene names | APOL2 | |||
|
Domain Architecture |
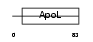
|
|||||
| Description | Apolipoprotein-L2 (Apolipoprotein L-II) (ApoL-II). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.009522 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
TNNT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.029123 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.060304 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
HS70B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70A_MOUSE
|
||||||
| NC score | 0.999929 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HSP71_HUMAN
|
||||||
| NC score | 0.998510 (rank : 3) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HS70L_MOUSE
|
||||||
| NC score | 0.997594 (rank : 4) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HS70L_HUMAN
|
||||||
| NC score | 0.997341 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HSP72_HUMAN
|
||||||
| NC score | 0.996792 (rank : 6) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP72_MOUSE
|
||||||
| NC score | 0.996529 (rank : 7) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP76_HUMAN
|
||||||
| NC score | 0.994867 (rank : 8) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP7C_HUMAN
|
||||||
| NC score | 0.993321 (rank : 9) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| NC score | 0.992668 (rank : 10) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP78_HUMAN
|
||||||
| NC score | 0.983557 (rank : 11) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| NC score | 0.983454 (rank : 12) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.982710 (rank : 13) | θ value | 2.77624e-142 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.981857 (rank : 14) | θ value | 1.92261e-143 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
STCH_HUMAN
|
||||||
| NC score | 0.963460 (rank : 15) | θ value | 2.09745e-73 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_MOUSE
|
||||||
| NC score | 0.963131 (rank : 16) | θ value | 1.35953e-72 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS105_MOUSE
|
||||||
| NC score | 0.905607 (rank : 17) | θ value | 1.96316e-71 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
HS74L_MOUSE
|
||||||
| NC score | 0.902734 (rank : 18) | θ value | 2.92676e-75 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HSP74_HUMAN
|
||||||
| NC score | 0.902604 (rank : 19) | θ value | 1.45253e-74 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
HS74L_HUMAN
|
||||||
| NC score | 0.901311 (rank : 20) | θ value | 1.22964e-73 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.898144 (rank : 21) | θ value | 1.50314e-71 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.891054 (rank : 22) | θ value | 8.51556e-75 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.874423 (rank : 23) | θ value | 9.16162e-53 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| NC score | 0.253812 (rank : 24) | θ value | 5.44631e-05 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
ANR45_MOUSE
|
||||||
| NC score | 0.194545 (rank : 25) | θ value | 0.0148317 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_MOUSE
|
||||||
| NC score | 0.144997 (rank : 26) | θ value | 0.00298849 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
HS12B_HUMAN
|
||||||
| NC score | 0.090995 (rank : 27) | θ value | 0.0563607 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
PCIF1_HUMAN
|
||||||
| NC score | 0.065451 (rank : 28) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
COVA1_HUMAN
|
||||||
| NC score | 0.062069 (rank : 29) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.060304 (rank : 30) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.057481 (rank : 31) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
PCIF1_MOUSE
|
||||||
| NC score | 0.057120 (rank : 32) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59114 | Gene names | Pcif1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.056045 (rank : 33) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
COVA1_MOUSE
|
||||||
| NC score | 0.050289 (rank : 34) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.046249 (rank : 35) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.045496 (rank : 36) | θ value | 0.0330416 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.041816 (rank : 37) | θ value | 0.0563607 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.041469 (rank : 38) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.041431 (rank : 39) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.040984 (rank : 40) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.039534 (rank : 41) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.038383 (rank : 42) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
UBXD2_HUMAN
|
||||||
| NC score | 0.034722 (rank : 43) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.033359 (rank : 44) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.033275 (rank : 45) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
UBXD2_MOUSE
|
||||||
| NC score | 0.032176 (rank : 46) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.031004 (rank : 47) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.029949 (rank : 48) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.029947 (rank : 49) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
TNNT2_HUMAN
|
||||||
| NC score | 0.029123 (rank : 50) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.029023 (rank : 51) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.027888 (rank : 52) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.027816 (rank : 53) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.027506 (rank : 54) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.026296 (rank : 55) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.025871 (rank : 56) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.025338 (rank : 57) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
FLNA_MOUSE
|
||||||
| NC score | 0.023324 (rank : 58) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.023035 (rank : 59) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.022104 (rank : 60) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.021836 (rank : 61) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
PDP1_HUMAN
|
||||||
| NC score | 0.021063 (rank : 62) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0J1, Q5U5K1 | Gene names | PDP1, PDP, PPM2C | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
PDP1_MOUSE
|
||||||
| NC score | 0.020982 (rank : 63) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UV70 | Gene names | Pdp1, Ppm2c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 1, mitochondrial precursor (EC 3.1.3.43) (PDP 1) (Pyruvate dehydrogenase phosphatase, catalytic subunit 1) (PDPC 1) (Protein phosphatase 2C). | |||||
|
DZIP3_MOUSE
|
||||||
| NC score | 0.018808 (rank : 64) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
K2C3_HUMAN
|
||||||
| NC score | 0.018732 (rank : 65) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.018510 (rank : 66) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.017778 (rank : 67) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.016464 (rank : 68) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
K2C1_HUMAN
|
||||||
| NC score | 0.011936 (rank : 69) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
K2C7_HUMAN
|
||||||
| NC score | 0.011916 (rank : 70) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08729, Q92676, Q9BUD8, Q9Y3R7 | Gene names | KRT7, SCL | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin). | |||||
|
APOL2_HUMAN
|
||||||
| NC score | 0.010027 (rank : 71) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQE5, O95915, Q5TH96, Q969T6, Q9BT28, Q9UGT1, Q9UH10 | Gene names | APOL2 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein-L2 (Apolipoprotein L-II) (ApoL-II). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.009522 (rank : 72) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
E2AK2_MOUSE
|
||||||
| NC score | 0.007519 (rank : 73) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||