Please be patient as the page loads
|
HSP72_MOUSE
|
||||||
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GRP78_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.983285 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.983268 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
HS70A_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.996492 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.996529 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70L_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.996457 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HS70L_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.996856 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HSP71_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.996185 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HSP72_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.999624 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP72_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP76_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.993691 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP7C_HUMAN
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.993480 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.993227 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 3.50381e-145 (rank : 13) | NC score | 0.981658 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 5.9766e-145 (rank : 14) | NC score | 0.982851 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
STCH_HUMAN
|
||||||
| θ value | 7.97034e-73 (rank : 15) | NC score | 0.964290 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_MOUSE
|
||||||
| θ value | 2.31901e-72 (rank : 16) | NC score | 0.963988 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS74L_MOUSE
|
||||||
| θ value | 9.74306e-71 (rank : 17) | NC score | 0.899733 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 2.3998e-69 (rank : 18) | NC score | 0.887294 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
HS74L_HUMAN
|
||||||
| θ value | 5.34618e-69 (rank : 19) | NC score | 0.897645 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HSP74_HUMAN
|
||||||
| θ value | 1.19101e-68 (rank : 20) | NC score | 0.899772 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
HS105_HUMAN
|
||||||
| θ value | 2.24614e-67 (rank : 21) | NC score | 0.895936 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HS105_MOUSE
|
||||||
| θ value | 6.53529e-67 (rank : 22) | NC score | 0.902890 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 2.25659e-51 (rank : 23) | NC score | 0.871828 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 24) | NC score | 0.254361 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 25) | NC score | 0.077145 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
ANR45_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.195566 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.030796 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.023839 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.015757 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.066691 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.050002 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.035175 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
PARG_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.024816 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.022488 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
FLNA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.029965 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.022859 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.037291 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.032312 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.030697 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PABP4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.012782 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.023514 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.018179 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.034223 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
HS12B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.125555 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.041734 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
K2C3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.016453 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |
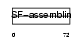
|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.046898 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PCIF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.053481 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.028820 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
COVA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.060047 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |
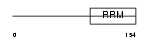
|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
COVA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.049114 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.055967 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.026399 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
PCIF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.045046 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59114 | Gene names | Pcif1 | |||
|
Domain Architecture |
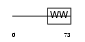
|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.024918 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
SYWM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.038379 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYK1 | Gene names | Wars2 | |||
|
Domain Architecture |
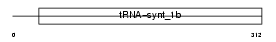
|
|||||
| Description | Tryptophanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) ((Mt)TrpRS). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.019549 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.011589 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.014522 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.025231 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.018865 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.015836 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.019465 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.015044 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.012452 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
AVIL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.003904 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75366 | Gene names | AVIL | |||
|
Domain Architecture |

|
|||||
| Description | Advillin (p92). | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.022717 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | -0.001014 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.013106 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
CENPU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.040114 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
CLUS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.015814 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10909, P11380, P11381, Q7Z5B9 | Gene names | CLU | |||
|
Domain Architecture |

|
|||||
| Description | Clusterin precursor (Complement-associated protein SP-40,40) (Complement cytolysis inhibitor) (CLI) (NA1/NA2) (Apolipoprotein J) (Apo-J) (Testosterone-repressed prostate message 2) (TRPM-2) [Contains: Clusterin beta chain (ApoJalpha) (Complement cytolysis inhibitor a chain); Clusterin alpha chain (ApoJbeta) (Complement cytolysis inhibitor b chain)]. | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.006478 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
K2C1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.009771 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.018576 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.019717 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.004525 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
PABP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.004314 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.004288 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PCDH7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | -0.000374 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60245, O60246, O60247 | Gene names | PCDH7, BHPCDH | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-7 precursor (Brain-heart protocadherin) (BH-Pcdh). | |||||
|
PESC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.007439 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.019315 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
REST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.022187 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.024034 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.053622 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
HS12B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.071240 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
HSP72_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP72_HUMAN
|
||||||
| NC score | 0.999624 (rank : 2) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HS70L_MOUSE
|
||||||
| NC score | 0.996856 (rank : 3) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HS70B_MOUSE
|
||||||
| NC score | 0.996529 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70A_MOUSE
|
||||||
| NC score | 0.996492 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70L_HUMAN
|
||||||
| NC score | 0.996457 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HSP71_HUMAN
|
||||||
| NC score | 0.996185 (rank : 7) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HSP76_HUMAN
|
||||||
| NC score | 0.993691 (rank : 8) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP7C_HUMAN
|
||||||
| NC score | 0.993480 (rank : 9) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| NC score | 0.993227 (rank : 10) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP78_HUMAN
|
||||||
| NC score | 0.983285 (rank : 11) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| NC score | 0.983268 (rank : 12) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.982851 (rank : 13) | θ value | 5.9766e-145 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.981658 (rank : 14) | θ value | 3.50381e-145 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
STCH_HUMAN
|
||||||
| NC score | 0.964290 (rank : 15) | θ value | 7.97034e-73 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_MOUSE
|
||||||
| NC score | 0.963988 (rank : 16) | θ value | 2.31901e-72 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS105_MOUSE
|
||||||
| NC score | 0.902890 (rank : 17) | θ value | 6.53529e-67 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
HSP74_HUMAN
|
||||||
| NC score | 0.899772 (rank : 18) | θ value | 1.19101e-68 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
HS74L_MOUSE
|
||||||
| NC score | 0.899733 (rank : 19) | θ value | 9.74306e-71 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS74L_HUMAN
|
||||||
| NC score | 0.897645 (rank : 20) | θ value | 5.34618e-69 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.895936 (rank : 21) | θ value | 2.24614e-67 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.887294 (rank : 22) | θ value | 2.3998e-69 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.871828 (rank : 23) | θ value | 2.25659e-51 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| NC score | 0.254361 (rank : 24) | θ value | 0.000270298 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
ANR45_MOUSE
|
||||||
| NC score | 0.195566 (rank : 25) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_MOUSE
|
||||||
| NC score | 0.125555 (rank : 26) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.077145 (rank : 27) | θ value | 0.00509761 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
HS12B_HUMAN
|
||||||
| NC score | 0.071240 (rank : 28) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.066691 (rank : 29) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
COVA1_HUMAN
|
||||||
| NC score | 0.060047 (rank : 30) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.055967 (rank : 31) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.053622 (rank : 32) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
PCIF1_HUMAN
|
||||||
| NC score | 0.053481 (rank : 33) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.050002 (rank : 34) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
COVA1_MOUSE
|
||||||
| NC score | 0.049114 (rank : 35) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.046898 (rank : 36) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PCIF1_MOUSE
|
||||||
| NC score | 0.045046 (rank : 37) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59114 | Gene names | Pcif1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.041734 (rank : 38) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
CENPU_HUMAN
|
||||||
| NC score | 0.040114 (rank : 39) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
SYWM_MOUSE
|
||||||
| NC score | 0.038379 (rank : 40) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYK1 | Gene names | Wars2 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptophanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) ((Mt)TrpRS). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.037291 (rank : 41) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.035175 (rank : 42) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.034223 (rank : 43) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.032312 (rank : 44) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.030796 (rank : 45) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.030697 (rank : 46) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
FLNA_MOUSE
|
||||||
| NC score | 0.029965 (rank : 47) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.028820 (rank : 48) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.026399 (rank : 49) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.025231 (rank : 50) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.024918 (rank : 51) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
PARG_HUMAN
|
||||||
| NC score | 0.024816 (rank : 52) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.024034 (rank : 53) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.023839 (rank : 54) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.023514 (rank : 55) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.022859 (rank : 56) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.022717 (rank : 57) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.022488 (rank : 58) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.022187 (rank : 59) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.019717 (rank : 60) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.019549 (rank : 61) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.019465 (rank : 62) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.019315 (rank : 63) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.018865 (rank : 64) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.018576 (rank : 65) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.018179 (rank : 66) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
K2C3_HUMAN
|
||||||
| NC score | 0.016453 (rank : 67) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.015836 (rank : 68) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
CLUS_HUMAN
|
||||||
| NC score | 0.015814 (rank : 69) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10909, P11380, P11381, Q7Z5B9 | Gene names | CLU | |||
|
Domain Architecture |

|
|||||
| Description | Clusterin precursor (Complement-associated protein SP-40,40) (Complement cytolysis inhibitor) (CLI) (NA1/NA2) (Apolipoprotein J) (Apo-J) (Testosterone-repressed prostate message 2) (TRPM-2) [Contains: Clusterin beta chain (ApoJalpha) (Complement cytolysis inhibitor a chain); Clusterin alpha chain (ApoJbeta) (Complement cytolysis inhibitor b chain)]. | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.015757 (rank : 70) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.015044 (rank : 71) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.014522 (rank : 72) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.013106 (rank : 73) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
PABP4_HUMAN
|
||||||
| NC score | 0.012782 (rank : 74) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.012452 (rank : 75) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.011589 (rank : 76) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
K2C1_HUMAN
|
||||||
| NC score | 0.009771 (rank : 77) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
PESC_MOUSE
|
||||||
| NC score | 0.007439 (rank : 78) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.006478 (rank : 79) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.004525 (rank : 80) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
PABP1_HUMAN
|
||||||
| NC score | 0.004314 (rank : 81) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11940, Q15097, Q93004 | Gene names | PABPC1, PAB1, PABP1, PABPC2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
PABP1_MOUSE
|
||||||
| NC score | 0.004288 (rank : 82) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29341 | Gene names | Pabpc1, Pabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 1 (Poly(A)-binding protein 1) (PABP 1). | |||||
|
AVIL_HUMAN
|
||||||
| NC score | 0.003904 (rank : 83) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75366 | Gene names | AVIL | |||
|
Domain Architecture |

|
|||||
| Description | Advillin (p92). | |||||
|
PCDH7_HUMAN
|
||||||
| NC score | -0.000374 (rank : 84) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60245, O60246, O60247 | Gene names | PCDH7, BHPCDH | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-7 precursor (Brain-heart protocadherin) (BH-Pcdh). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | -0.001014 (rank : 85) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||