Please be patient as the page loads
|
PESC_MOUSE
|
||||||
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PESC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.958089 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00541, Q6IC29 | Gene names | PES1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
PESC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 3) | NC score | 0.092054 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 4) | NC score | 0.060864 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.131037 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.087482 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.144291 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.120493 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.085456 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
RADI_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.063698 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RADI_MOUSE
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.064550 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.048838 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
GAS8_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.083772 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.049517 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
XKR9_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.089901 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH62 | Gene names | Xkr9, Gm1620, Xrg9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 9. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.088531 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.062701 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.083837 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
LRC59_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.047336 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q922Q8, Q3TJ35, Q3TLC7, Q3TWT9, Q3TX86 | Gene names | Lrrc59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
XE7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.084753 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
XRCC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.110061 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
LRC59_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.046591 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.055135 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NDE1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.069911 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.071848 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
XKR9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.082217 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH70 | Gene names | XKR9, XRG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 9. | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.051638 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.073763 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CF060_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.066598 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.108838 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.101116 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.064064 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SEPT6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.023104 (rank : 150) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14141, Q5JTK0, Q969W5, Q96A13, Q96GR1, Q96P86, Q96P87 | Gene names | SEPT6, KIAA0128, SEP2 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-6. | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.048883 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CN140_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.073047 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CCG1, Q8CCM8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf140 homolog. | |||||
|
GAS8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.071886 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.088695 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.036225 (rank : 133) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
BLMH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.038769 (rank : 128) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13867, Q53F86, Q9UER9 | Gene names | BLMH | |||
|
Domain Architecture |
|
|||||
| Description | Bleomycin hydrolase (EC 3.4.22.40) (BLM hydrolase) (BMH) (BH). | |||||
|
CCD37_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.068717 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q494V2, Q494V1, Q494V4, Q8N838 | Gene names | CCDC37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
FRM4B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.040389 (rank : 126) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
MYO6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.037560 (rank : 131) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
TNNT3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.055155 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.035935 (rank : 135) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.076628 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.063441 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.028858 (rank : 141) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
NDE1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.068182 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
CN037_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.036108 (rank : 134) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.046499 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.042810 (rank : 122) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.050708 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CDC37_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.066092 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |
|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
DKC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.029627 (rank : 139) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60832, O43845, Q96G67, Q9Y505 | Gene names | DKC1, NOLA4 | |||
|
Domain Architecture |
|
|||||
| Description | H/ACA ribonucleoprotein complex subunit 4 (EC 5.4.99.-) (Dyskerin) (Nucleolar protein family A member 4) (snoRNP protein DKC1) (Nopp140- associated protein of 57 kDa) (Nucleolar protein NAP57) (CBF5 homolog). | |||||
|
FRM4B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.036754 (rank : 132) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
MX2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.022733 (rank : 151) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
NUCB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.039190 (rank : 127) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.057231 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
TSH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.032285 (rank : 138) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
UT14B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.053250 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.008324 (rank : 162) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.071672 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAN6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.015981 (rank : 157) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.052387 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.033867 (rank : 136) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MPP10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.057792 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.041408 (rank : 123) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.018018 (rank : 155) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.061361 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.041383 (rank : 124) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.056792 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.050233 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
IPP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.028903 (rank : 140) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.042954 (rank : 121) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.062855 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.026389 (rank : 145) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MUCB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.012459 (rank : 161) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04220 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig mu heavy chain disease protein (BOT). | |||||
|
PA24A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.012586 (rank : 160) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
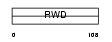
RWDD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.024046 (rank : 148) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQK7 | Gene names | Rwdd1 | |||
|
Domain Architecture |
|
|||||
| Description | RWD domain-containing protein 1 (IH1). | |||||
|
SH24A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.047530 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
ZFY19_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.020921 (rank : 153) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.070940 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
EGR4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.000815 (rank : 165) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.025235 (rank : 147) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.049750 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
LIPB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.041347 (rank : 125) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.045314 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.038209 (rank : 129) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
NOP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.021269 (rank : 152) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2X3, Q6PK08, Q9P036, Q9UFN3 | Gene names | NOP5 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein NOP5 (Nucleolar protein 5) (NOP58). | |||||
|
OTUD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.026860 (rank : 144) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T2D3, O75047 | Gene names | OTUD3, KIAA0459 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 3. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.047540 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
PRDM5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | -0.000754 (rank : 168) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CXE0 | Gene names | Prdm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.013812 (rank : 159) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.019872 (rank : 154) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.037898 (rank : 130) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.028269 (rank : 143) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.028799 (rank : 142) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.067083 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.008059 (rank : 163) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.048905 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
HSP72_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.007439 (rank : 164) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.033470 (rank : 137) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.016054 (rank : 156) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.015957 (rank : 158) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.044346 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.026078 (rank : 146) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
PTCA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.023152 (rank : 149) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64697 | Gene names | Ptprcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase receptor type C-associated protein (PTPRC-associated protein) (CD45-associated protein) (CD45-AP) (LSM- 1). | |||||
|
STK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.000153 (rank : 167) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JI11 | Gene names | Stk4, Mst1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 4 (EC 2.7.11.1) (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1). | |||||
|
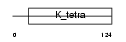
ZBTB6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.000397 (rank : 166) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051301 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.051483 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.061196 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CCD19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.054796 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.054364 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.052226 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCD91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.056825 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CCD91_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.051574 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.052687 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CDC37_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051882 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
CI039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051076 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CI093_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.052042 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CING_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.050471 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CING_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.052295 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.063135 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.053468 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.053931 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.060436 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.055896 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.057672 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.054128 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.071804 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.053197 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.065876 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.065521 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
K0555_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.052326 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.050622 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
KTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.051730 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LRRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.065281 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.050527 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.050352 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MOES_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.057498 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MOES_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.057117 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.051246 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.052665 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.050322 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
NDEL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.052926 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
NDEL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.052804 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.051220 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PININ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051115 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.058190 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.053377 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050156 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050141 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.066614 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.056131 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.051888 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.051434 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.054191 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.053248 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.050795 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.050361 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.050664 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.062464 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.050421 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.050347 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.054897 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.051888 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.051413 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
PESC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
PESC_HUMAN
|
||||||
| NC score | 0.958089 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00541, Q6IC29 | Gene names | PES1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.144291 (rank : 3) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.131037 (rank : 4) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.120493 (rank : 5) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
XRCC1_HUMAN
|
||||||
| NC score | 0.110061 (rank : 6) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18887, Q9HCB1 | Gene names | XRCC1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.108838 (rank : 7) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.101116 (rank : 8) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.092054 (rank : 9) | θ value | 0.00228821 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
XKR9_MOUSE
|
||||||
| NC score | 0.089901 (rank : 10) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH62 | Gene names | Xkr9, Gm1620, Xrg9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 9. | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.088695 (rank : 11) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.088531 (rank : 12) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.087482 (rank : 13) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.085456 (rank : 14) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.084753 (rank : 15) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.083837 (rank : 16) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
GAS8_MOUSE
|
||||||
| NC score | 0.083772 (rank : 17) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
XKR9_HUMAN
|
||||||
| NC score | 0.082217 (rank : 18) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5GH70 | Gene names | XKR9, XRG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 9. | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.076628 (rank : 19) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.073763 (rank : 20) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CN140_MOUSE
|
||||||
| NC score | 0.073047 (rank : 21) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CCG1, Q8CCM8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf140 homolog. | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.071886 (rank : 22) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.071848 (rank : 23) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.071804 (rank : 24) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.071672 (rank : 25) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.070940 (rank : 26) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.069911 (rank : 27) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
CCD37_HUMAN
|
||||||
| NC score | 0.068717 (rank : 28) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q494V2, Q494V1, Q494V4, Q8N838 | Gene names | CCDC37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
NDE1_MOUSE
|
||||||
| NC score | 0.068182 (rank : 29) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.067083 (rank : 30) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.066614 (rank : 31) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.066598 (rank : 32) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CDC37_MOUSE
|
||||||
| NC score | 0.066092 (rank : 33) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.065876 (rank : 34) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.065521 (rank : 35) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
LRRF2_MOUSE
|
||||||
| NC score | 0.065281 (rank : 36) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.064550 (rank : 37) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.064064 (rank : 38) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.063698 (rank : 39) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.063441 (rank : 40) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.063135 (rank : 41) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.062855 (rank : 42) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.062701 (rank : 43) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.062464 (rank : 44) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.061361 (rank : 45) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.061196 (rank : 46) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.060864 (rank : 47) | θ value | 0.00869519 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.060436 (rank : 48) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.058190 (rank : 49) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
MPP10_HUMAN
|
||||||
| NC score | 0.057792 (rank : 50) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.057672 (rank : 51) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.057498 (rank : 52) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.057231 (rank : 53) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.057117 (rank : 54) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.056825 (rank : 55) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.056792 (rank : 56) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.056131 (rank : 57) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.055896 (rank : 58) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
TNNT3_MOUSE
|
||||||
| NC score | 0.055155 (rank : 59) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.055135 (rank : 60) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.054897 (rank : 61) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CCD19_HUMAN
|
||||||
| NC score | 0.054796 (rank : 62) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.054364 (rank : 63) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.054191 (rank : 64) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.054128 (rank : 65) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.053931 (rank : 66) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.053468 (rank : 67) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.053377 (rank : 68) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
UT14B_MOUSE
|
||||||
| NC score | 0.053250 (rank : 69) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.053248 (rank : 70) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.053197 (rank : 71) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NDEL1_HUMAN
|
||||||
| NC score | 0.052926 (rank : 72) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9GZM8, Q86T80, Q8TAR7, Q9UH50 | Gene names | NDEL1, EOPA, MITAP1, NUDEL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Mitosin- associated protein 1). | |||||
|
NDEL1_MOUSE
|
||||||
| NC score | 0.052804 (rank : 73) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ERR1, Q9D0Q4, Q9EPT6 | Gene names | Ndel1, Nudel | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE-like 1 (Protein Nudel) (Protein mNudE-like) (mNudE-L). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.052687 (rank : 74) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.052665 (rank : 75) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.052387 (rank : 76) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.052326 (rank : 77) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.052295 (rank : 78) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.052226 (rank : 79) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.052042 (rank : 80) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.051888 (rank : 81) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.051888 (rank : 82) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CDC37_HUMAN
|
||||||
| NC score | 0.051882 (rank : 83) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.051730 (rank : 84) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.051638 (rank : 85) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CCD91_MOUSE
|
||||||
| NC score | 0.051574 (rank : 86) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.051483 (rank : 87) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.051434 (rank : 88) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.051413 (rank : 89) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.051301 (rank : 90) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.051246 (rank : 91) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.051220 (rank : 92) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.051115 (rank : 93) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.051076 (rank : 94) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.050795 (rank : 95) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.050708 (rank : 96) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.050664 (rank : 97) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.050622 (rank : 98) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.050527 (rank : 99) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.050471 (rank : 100) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.050421 (rank : 101) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.050361 (rank : 102) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.050352 (rank : 103) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.050347 (rank : 104) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.050322 (rank : 105) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.050233 (rank : 106) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.050156 (rank : 107) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.050141 (rank : 108) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.049750 (rank : 109) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.049517 (rank : 110) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.048905 (rank : 111) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.048883 (rank : 112) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.048838 (rank : 113) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.047540 (rank : 114) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SH24A_HUMAN
|
||||||
| NC score | 0.047530 (rank : 115) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
LRC59_MOUSE
|
||||||
| NC score | 0.047336 (rank : 116) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q922Q8, Q3TJ35, Q3TLC7, Q3TWT9, Q3TX86 | Gene names | Lrrc59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
LRC59_HUMAN
|
||||||
| NC score | 0.046591 (rank : 117) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.046499 (rank : 118) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.045314 (rank : 119) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.044346 (rank : 120) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.042954 (rank : 121) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.042810 (rank : 122) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.041408 (rank : 123) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.041383 (rank : 124) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
LIPB1_MOUSE
|
||||||
| NC score | 0.041347 (rank : 125) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
FRM4B_HUMAN
|
||||||
| NC score | 0.040389 (rank : 126) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2L6, Q8TAI3 | Gene names | FRMD4B, KIAA1013 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1). | |||||
|
NUCB2_MOUSE
|
||||||
| NC score | 0.039190 (rank : 127) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
BLMH_HUMAN
|
||||||
| NC score | 0.038769 (rank : 128) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13867, Q53F86, Q9UER9 | Gene names | BLMH | |||
|
Domain Architecture |
|
|||||
| Description | Bleomycin hydrolase (EC 3.4.22.40) (BLM hydrolase) (BMH) (BH). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.038209 (rank : 129) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.037898 (rank : 130) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MYO6_MOUSE
|
||||||
| NC score | 0.037560 (rank : 131) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
FRM4B_MOUSE
|
||||||
| NC score | 0.036754 (rank : 132) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q920B0, Q3V1S1, Q9ESP9 | Gene names | Frmd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM domain-containing protein 4B (GRP1-binding protein GRSP1) (Golgi- associated band 4.1-like protein) (GOBLIN). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.036225 (rank : 133) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CN037_HUMAN
|
||||||
| NC score | 0.036108 (rank : 134) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.035935 (rank : 135) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.033867 (rank : 136) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.033470 (rank : 137) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
TSH2_HUMAN
|
||||||
| NC score | 0.032285 (rank : 138) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
DKC1_HUMAN
|
||||||
| NC score | 0.029627 (rank : 139) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60832, O43845, Q96G67, Q9Y505 | Gene names | DKC1, NOLA4 | |||
|
Domain Architecture |

|
|||||
| Description | H/ACA ribonucleoprotein complex subunit 4 (EC 5.4.99.-) (Dyskerin) (Nucleolar protein family A member 4) (snoRNP protein DKC1) (Nopp140- associated protein of 57 kDa) (Nucleolar protein NAP57) (CBF5 homolog). | |||||
|
IPP4_HUMAN
|
||||||
| NC score | 0.028903 (rank : 140) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.028858 (rank : 141) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.028799 (rank : 142) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.028269 (rank : 143) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
OTUD3_HUMAN
|
||||||
| NC score | 0.026860 (rank : 144) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T2D3, O75047 | Gene names | OTUD3, KIAA0459 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 3. | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.026389 (rank : 145) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.026078 (rank : 146) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.025235 (rank : 147) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
RWDD1_MOUSE
|
||||||
| NC score | 0.024046 (rank : 148) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQK7 | Gene names | Rwdd1 | |||
|
Domain Architecture |

|
|||||
| Description | RWD domain-containing protein 1 (IH1). | |||||
|
PTCA_MOUSE
|
||||||
| NC score | 0.023152 (rank : 149) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64697 | Gene names | Ptprcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase receptor type C-associated protein (PTPRC-associated protein) (CD45-associated protein) (CD45-AP) (LSM- 1). | |||||
|
SEPT6_HUMAN
|
||||||
| NC score | 0.023104 (rank : 150) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14141, Q5JTK0, Q969W5, Q96A13, Q96GR1, Q96P86, Q96P87 | Gene names | SEPT6, KIAA0128, SEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-6. | |||||
|
MX2_MOUSE
|
||||||
| NC score | 0.022733 (rank : 151) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVP9, Q922L4 | Gene names | Mx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced GTP-binding protein Mx2. | |||||
|
NOP5_HUMAN
|
||||||
| NC score | 0.021269 (rank : 152) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2X3, Q6PK08, Q9P036, Q9UFN3 | Gene names | NOP5 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein NOP5 (Nucleolar protein 5) (NOP58). | |||||
|
ZFY19_HUMAN
|
||||||
| NC score | 0.020921 (rank : 153) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.019872 (rank : 154) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.018018 (rank : 155) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.016054 (rank : 156) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
CAN6_HUMAN
|
||||||
| NC score | 0.015981 (rank : 157) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6Q1, Q9UEQ1, Q9UJA8 | Gene names | CAPN6, CALPM, CANPX | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-6 (Calpamodulin) (CalpM) (Calpain-like protease X-linked). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.015957 (rank : 158) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.013812 (rank : 159) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
PA24A_MOUSE
|
||||||
| NC score | 0.012586 (rank : 160) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
MUCB_HUMAN
|
||||||
| NC score | 0.012459 (rank : 161) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04220 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig mu heavy chain disease protein (BOT). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.008324 (rank : 162) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.008059 (rank : 163) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
HSP72_MOUSE
|
||||||
| NC score | 0.007439 (rank : 164) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
EGR4_MOUSE
|
||||||
| NC score | 0.000815 (rank : 165) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||
|
ZBTB6_HUMAN
|
||||||
| NC score | 0.000397 (rank : 166) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||
|
STK4_MOUSE
|
||||||
| NC score | 0.000153 (rank : 167) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JI11 | Gene names | Stk4, Mst1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 4 (EC 2.7.11.1) (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1). | |||||
|
PRDM5_MOUSE
|
||||||
| NC score | -0.000754 (rank : 168) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CXE0 | Gene names | Prdm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 5 (PR domain-containing protein 5). | |||||