Please be patient as the page loads
|
HS70L_MOUSE
|
||||||
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GRP78_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.982379 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982323 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
HS70A_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.997560 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.997594 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70L_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999079 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HS70L_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HSP71_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.997053 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HSP72_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.996677 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP72_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.996856 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP76_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.994269 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP7C_HUMAN
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.992299 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.991551 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 2.19465e-147 (rank : 13) | NC score | 0.983547 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 6.38546e-147 (rank : 14) | NC score | 0.984746 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
HS74L_MOUSE
|
||||||
| θ value | 4.99229e-75 (rank : 15) | NC score | 0.903843 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS74L_HUMAN
|
||||||
| θ value | 1.89707e-74 (rank : 16) | NC score | 0.901134 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 4.22625e-74 (rank : 17) | NC score | 0.890200 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
HSP74_HUMAN
|
||||||
| θ value | 1.22964e-73 (rank : 18) | NC score | 0.903799 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
STCH_HUMAN
|
||||||
| θ value | 4.67263e-73 (rank : 19) | NC score | 0.965519 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS105_HUMAN
|
||||||
| θ value | 6.10265e-73 (rank : 20) | NC score | 0.899562 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HS105_MOUSE
|
||||||
| θ value | 5.1662e-72 (rank : 21) | NC score | 0.907081 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
STCH_MOUSE
|
||||||
| θ value | 8.81223e-72 (rank : 22) | NC score | 0.965174 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 2.25659e-51 (rank : 23) | NC score | 0.873594 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 24) | NC score | 0.249394 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 25) | NC score | 0.143839 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
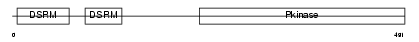
E2AK2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.008463 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
ANR45_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.190380 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.089638 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.024836 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
CELR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.002447 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.008555 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.021814 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
K2C3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.014934 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
PWP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.010799 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LL5, Q9D6T6 | Gene names | Pwp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periodic tryptophan protein 1 homolog. | |||||
|
SYWM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.038147 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYK1 | Gene names | Wars2 | |||
|
Domain Architecture |
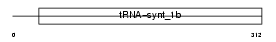
|
|||||
| Description | Tryptophanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) ((Mt)TrpRS). | |||||
|
K2C1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.006332 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
UBXD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.031329 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
UBXD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.029967 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
SLIT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.001508 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75094, O95804, Q9UFH5 | Gene names | SLIT3, KIAA0814, MEGF5, SLIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Multiple epidermal growth factor-like domains 5). | |||||
|
SLIT3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.002061 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WVB4 | Gene names | Slit3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Slit3). | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.001242 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.051465 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
K2C4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.005854 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
TTC12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.003373 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.018385 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.023019 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
GPR98_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.003175 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.000484 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
K2C7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.008093 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08729, Q92676, Q9BUD8, Q9Y3R7 | Gene names | KRT7, SCL | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.057500 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
HS70L_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HS70L_HUMAN
|
||||||
| NC score | 0.999079 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HS70B_MOUSE
|
||||||
| NC score | 0.997594 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70A_MOUSE
|
||||||
| NC score | 0.997560 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HSP71_HUMAN
|
||||||
| NC score | 0.997053 (rank : 5) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HSP72_MOUSE
|
||||||
| NC score | 0.996856 (rank : 6) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP72_HUMAN
|
||||||
| NC score | 0.996677 (rank : 7) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP76_HUMAN
|
||||||
| NC score | 0.994269 (rank : 8) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP7C_HUMAN
|
||||||
| NC score | 0.992299 (rank : 9) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| NC score | 0.991551 (rank : 10) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.984746 (rank : 11) | θ value | 6.38546e-147 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.983547 (rank : 12) | θ value | 2.19465e-147 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP78_HUMAN
|
||||||
| NC score | 0.982379 (rank : 13) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| NC score | 0.982323 (rank : 14) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
STCH_HUMAN
|
||||||
| NC score | 0.965519 (rank : 15) | θ value | 4.67263e-73 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_MOUSE
|
||||||
| NC score | 0.965174 (rank : 16) | θ value | 8.81223e-72 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS105_MOUSE
|
||||||
| NC score | 0.907081 (rank : 17) | θ value | 5.1662e-72 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
HS74L_MOUSE
|
||||||
| NC score | 0.903843 (rank : 18) | θ value | 4.99229e-75 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HSP74_HUMAN
|
||||||
| NC score | 0.903799 (rank : 19) | θ value | 1.22964e-73 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
HS74L_HUMAN
|
||||||
| NC score | 0.901134 (rank : 20) | θ value | 1.89707e-74 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.899562 (rank : 21) | θ value | 6.10265e-73 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.890200 (rank : 22) | θ value | 4.22625e-74 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.873594 (rank : 23) | θ value | 2.25659e-51 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| NC score | 0.249394 (rank : 24) | θ value | 0.00134147 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
ANR45_MOUSE
|
||||||
| NC score | 0.190380 (rank : 25) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_MOUSE
|
||||||
| NC score | 0.143839 (rank : 26) | θ value | 0.00298849 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
HS12B_HUMAN
|
||||||
| NC score | 0.089638 (rank : 27) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.057500 (rank : 28) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.051465 (rank : 29) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
SYWM_MOUSE
|
||||||
| NC score | 0.038147 (rank : 30) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYK1 | Gene names | Wars2 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptophanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) ((Mt)TrpRS). | |||||
|
UBXD2_HUMAN
|
||||||
| NC score | 0.031329 (rank : 31) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
UBXD2_MOUSE
|
||||||
| NC score | 0.029967 (rank : 32) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.024836 (rank : 33) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.023019 (rank : 34) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.021814 (rank : 35) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.018385 (rank : 36) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
K2C3_HUMAN
|
||||||
| NC score | 0.014934 (rank : 37) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
PWP1_MOUSE
|
||||||
| NC score | 0.010799 (rank : 38) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LL5, Q9D6T6 | Gene names | Pwp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periodic tryptophan protein 1 homolog. | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.008555 (rank : 39) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
E2AK2_MOUSE
|
||||||
| NC score | 0.008463 (rank : 40) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
K2C7_HUMAN
|
||||||
| NC score | 0.008093 (rank : 41) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08729, Q92676, Q9BUD8, Q9Y3R7 | Gene names | KRT7, SCL | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin). | |||||
|
K2C1_HUMAN
|
||||||
| NC score | 0.006332 (rank : 42) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
K2C4_MOUSE
|
||||||
| NC score | 0.005854 (rank : 43) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.003373 (rank : 44) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
GPR98_HUMAN
|
||||||
| NC score | 0.003175 (rank : 45) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
CELR1_MOUSE
|
||||||
| NC score | 0.002447 (rank : 46) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
SLIT3_MOUSE
|
||||||
| NC score | 0.002061 (rank : 47) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WVB4 | Gene names | Slit3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Slit3). | |||||
|
SLIT3_HUMAN
|
||||||
| NC score | 0.001508 (rank : 48) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75094, O95804, Q9UFH5 | Gene names | SLIT3, KIAA0814, MEGF5, SLIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Multiple epidermal growth factor-like domains 5). | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.001242 (rank : 49) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.000484 (rank : 50) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||