Please be patient as the page loads
|
HSP7C_MOUSE
|
||||||
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GRP78_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.983523 (rank : 11) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.983409 (rank : 12) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
HS70A_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.992631 (rank : 7) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.992668 (rank : 6) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70L_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.991959 (rank : 8) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HS70L_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.991551 (rank : 10) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HSP71_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.992828 (rank : 5) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HSP72_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.993613 (rank : 3) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP72_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.993227 (rank : 4) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP76_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.991899 (rank : 9) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP7C_HUMAN
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.999221 (rank : 2) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 146 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 166 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 1.68038e-147 (rank : 13) | NC score | 0.978184 (rank : 14) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 2.8663e-147 (rank : 14) | NC score | 0.978796 (rank : 13) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
HSP74_HUMAN
|
||||||
| θ value | 3.23591e-74 (rank : 15) | NC score | 0.900677 (rank : 18) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
STCH_HUMAN
|
||||||
| θ value | 3.23591e-74 (rank : 16) | NC score | 0.960252 (rank : 15) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 7.20884e-74 (rank : 17) | NC score | 0.890152 (rank : 22) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
STCH_MOUSE
|
||||||
| θ value | 1.60597e-73 (rank : 18) | NC score | 0.959044 (rank : 16) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS74L_MOUSE
|
||||||
| θ value | 3.57772e-73 (rank : 19) | NC score | 0.898926 (rank : 20) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS105_HUMAN
|
||||||
| θ value | 1.04096e-72 (rank : 20) | NC score | 0.895953 (rank : 21) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HS74L_HUMAN
|
||||||
| θ value | 1.7756e-72 (rank : 21) | NC score | 0.899703 (rank : 19) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS105_MOUSE
|
||||||
| θ value | 1.66191e-70 (rank : 22) | NC score | 0.902151 (rank : 17) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 2.66562e-52 (rank : 23) | NC score | 0.873748 (rank : 23) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 24) | NC score | 0.255652 (rank : 24) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
ANR45_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 25) | NC score | 0.196581 (rank : 25) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.049682 (rank : 78) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 27) | NC score | 0.058530 (rank : 43) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 28) | NC score | 0.090356 (rank : 27) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.071509 (rank : 31) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.056148 (rank : 48) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.075937 (rank : 29) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.053065 (rank : 62) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.038189 (rank : 112) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.055175 (rank : 52) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.066686 (rank : 35) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.065222 (rank : 37) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
REST_HUMAN
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.051445 (rank : 70) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
MACOI_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.049957 (rank : 76) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.064120 (rank : 38) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.032573 (rank : 125) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CING_MOUSE
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.047423 (rank : 93) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.038117 (rank : 114) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.067384 (rank : 33) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.066806 (rank : 34) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 45) | NC score | 0.047739 (rank : 91) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.048940 (rank : 82) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.039853 (rank : 111) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.062709 (rank : 39) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.066303 (rank : 36) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.030770 (rank : 129) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.048372 (rank : 89) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.034765 (rank : 122) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
CENPU_HUMAN
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.059697 (rank : 41) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
DEK_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.036094 (rank : 120) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
GBP2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.024763 (rank : 141) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z0E6, Q8CIC6, Q921N2, Q9R1I0 | Gene names | Gbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (mGBP2) (mGBP-2). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.056878 (rank : 45) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.055081 (rank : 53) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.047530 (rank : 92) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
WBP4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.030029 (rank : 130) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75554 | Gene names | WBP4, FBP21, FNBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.024437 (rank : 142) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
HS12B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.126611 (rank : 26) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.023584 (rank : 145) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
UT14B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.025971 (rank : 138) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.047825 (rank : 90) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.048410 (rank : 88) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.041576 (rank : 106) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.049871 (rank : 77) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.056242 (rank : 47) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.052451 (rank : 68) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.036901 (rank : 118) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.033534 (rank : 123) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.028526 (rank : 134) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.048438 (rank : 87) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.049255 (rank : 79) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.046287 (rank : 96) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.037978 (rank : 115) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.049167 (rank : 81) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 78) | NC score | 0.038145 (rank : 113) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.037475 (rank : 117) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 80) | NC score | 0.055846 (rank : 50) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.022136 (rank : 147) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.031819 (rank : 127) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.052464 (rank : 67) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.053140 (rank : 61) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.049249 (rank : 80) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.031077 (rank : 128) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.076006 (rank : 28) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.029957 (rank : 131) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.042220 (rank : 105) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
COVA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.069820 (rank : 32) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |
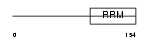
|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.055866 (rank : 49) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
FLNA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.025519 (rank : 139) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.058806 (rank : 42) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.053686 (rank : 58) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.056280 (rank : 46) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.048717 (rank : 86) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.048726 (rank : 85) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.046983 (rank : 95) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.052588 (rank : 66) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.062358 (rank : 40) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
IKKB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | -0.000534 (rank : 172) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14920, O75327 | Gene names | IKBKB, IKKB | |||
|
Domain Architecture |
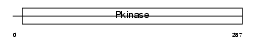
|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
IKKB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.000118 (rank : 171) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88351, Q9R1J6 | Gene names | Ikbkb, Ikkb | |||
|
Domain Architecture |
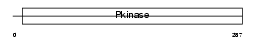
|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
K2C4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.020387 (rank : 149) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.057490 (rank : 44) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.052834 (rank : 65) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
PSIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.026448 (rank : 137) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
REST_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.053489 (rank : 59) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.047321 (rank : 94) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.023349 (rank : 146) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.055076 (rank : 54) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.048924 (rank : 83) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.051121 (rank : 71) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.051000 (rank : 72) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.025418 (rank : 140) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.042943 (rank : 104) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.050393 (rank : 74) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
CTGE5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.044898 (rank : 102) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
IRX5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.006839 (rank : 168) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKQ4, Q80WV4, Q9JLL5 | Gene names | Irx5, Irxb2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.054823 (rank : 55) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.052888 (rank : 64) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
ST18_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.019638 (rank : 152) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.040291 (rank : 109) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ARS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.018871 (rank : 153) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.053019 (rank : 63) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.029644 (rank : 132) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.028971 (rank : 133) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
FA40A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.007371 (rank : 167) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C079, Q80T92, Q8BZC6, Q8CIF7 | Gene names | Fam40a, Kiaa1761 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM40A. | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.048868 (rank : 84) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
HOME1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.044192 (rank : 103) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |
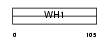
|
|||||
| Description | Homer protein homolog 1. | |||||
|
K2C3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.023851 (rank : 144) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.026817 (rank : 136) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.033426 (rank : 124) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PSA7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.015558 (rank : 156) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14818, Q9BR53, Q9H4K5, Q9UEU8 | Gene names | PSMA7 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 7 (EC 3.4.25.1) (Proteasome subunit RC6- 1) (Proteasome subunit XAPC7). | |||||
|
PSA7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.014128 (rank : 159) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2U0 | Gene names | Psma7 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 7 (EC 3.4.25.1) (Proteasome subunit RC6- 1). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.040262 (rank : 110) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.046193 (rank : 97) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.037481 (rank : 116) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CT116_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.027004 (rank : 135) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96HY6, Q9BW47 | Gene names | C20orf116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 precursor. | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.045732 (rank : 99) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.046184 (rank : 98) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.045384 (rank : 100) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.015753 (rank : 155) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
PABP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.008216 (rank : 166) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.020136 (rank : 150) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SGOL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.008734 (rank : 165) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CXH7, Q3U4K4, Q588H1, Q8BKW2 | Gene names | Sgol1, Sgo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1. | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.009017 (rank : 164) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.019840 (rank : 151) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TEX15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.014099 (rank : 160) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
TRML1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.011398 (rank : 163) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YW5, Q496B3, Q8IWY1, Q8IWY2 | Gene names | TREML1, TLT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 1 protein precursor (TLT-1) (Triggering receptor expressed on myeloid cells-like protein 1). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.036140 (rank : 119) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.015259 (rank : 157) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CCD38_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.015926 (rank : 154) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CDN8, Q8CDD6, Q8CDE1 | Gene names | Ccdc38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.001191 (rank : 170) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
KIF1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.001869 (rank : 169) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.020907 (rank : 148) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.024070 (rank : 143) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.040890 (rank : 108) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.051637 (rank : 69) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.053322 (rank : 60) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.015208 (rank : 158) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PCIF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.045325 (rank : 101) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.031849 (rank : 126) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
RWDD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.013326 (rank : 162) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H446, Q9Y313, Q9Y6B3 | Gene names | RWDD1 | |||
|
Domain Architecture |
|
|||||
| Description | RWD domain-containing protein 1. | |||||
|
RWDD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.013669 (rank : 161) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQK7 | Gene names | Rwdd1 | |||
|
Domain Architecture |
|
|||||
| Description | RWD domain-containing protein 1 (IH1). | |||||
|
SYWM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.035285 (rank : 121) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYK1 | Gene names | Wars2 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptophanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) ((Mt)TrpRS). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.041075 (rank : 107) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.050535 (rank : 73) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054248 (rank : 56) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
COVA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.053980 (rank : 57) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.055292 (rank : 51) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
HS12B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.072386 (rank : 30) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
MYH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.050307 (rank : 75) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
HSP7C_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 166 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_HUMAN
|
||||||
| NC score | 0.999221 (rank : 2) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 146 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP72_HUMAN
|
||||||
| NC score | 0.993613 (rank : 3) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP72_MOUSE
|
||||||
| NC score | 0.993227 (rank : 4) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP71_HUMAN
|
||||||
| NC score | 0.992828 (rank : 5) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HS70B_MOUSE
|
||||||
| NC score | 0.992668 (rank : 6) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70A_MOUSE
|
||||||
| NC score | 0.992631 (rank : 7) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70L_HUMAN
|
||||||
| NC score | 0.991959 (rank : 8) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HSP76_HUMAN
|
||||||
| NC score | 0.991899 (rank : 9) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HS70L_MOUSE
|
||||||
| NC score | 0.991551 (rank : 10) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
GRP78_HUMAN
|
||||||
| NC score | 0.983523 (rank : 11) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| NC score | 0.983409 (rank : 12) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.978796 (rank : 13) | θ value | 2.8663e-147 (rank : 14) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.978184 (rank : 14) | θ value | 1.68038e-147 (rank : 13) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
STCH_HUMAN
|
||||||
| NC score | 0.960252 (rank : 15) | θ value | 3.23591e-74 (rank : 16) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_MOUSE
|
||||||
| NC score | 0.959044 (rank : 16) | θ value | 1.60597e-73 (rank : 18) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS105_MOUSE
|
||||||
| NC score | 0.902151 (rank : 17) | θ value | 1.66191e-70 (rank : 22) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
HSP74_HUMAN
|
||||||
| NC score | 0.900677 (rank : 18) | θ value | 3.23591e-74 (rank : 15) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
HS74L_HUMAN
|
||||||
| NC score | 0.899703 (rank : 19) | θ value | 1.7756e-72 (rank : 21) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS74L_MOUSE
|
||||||
| NC score | 0.898926 (rank : 20) | θ value | 3.57772e-73 (rank : 19) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.895953 (rank : 21) | θ value | 1.04096e-72 (rank : 20) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.890152 (rank : 22) | θ value | 7.20884e-74 (rank : 17) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.873748 (rank : 23) | θ value | 2.66562e-52 (rank : 23) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| NC score | 0.255652 (rank : 24) | θ value | 5.44631e-05 (rank : 24) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
ANR45_MOUSE
|
||||||
| NC score | 0.196581 (rank : 25) | θ value | 0.00175202 (rank : 25) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_MOUSE
|
||||||
| NC score | 0.126611 (rank : 26) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.090356 (rank : 27) | θ value | 0.00869519 (rank : 28) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.076006 (rank : 28) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.075937 (rank : 29) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
HS12B_HUMAN
|
||||||
| NC score | 0.072386 (rank : 30) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.071509 (rank : 31) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 126 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
COVA1_HUMAN
|
||||||
| NC score | 0.069820 (rank : 32) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.067384 (rank : 33) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.066806 (rank : 34) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.066686 (rank : 35) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 129 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.066303 (rank : 36) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.065222 (rank : 37) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.064120 (rank : 38) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.062709 (rank : 39) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.062358 (rank : 40) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
CENPU_HUMAN
|
||||||
| NC score | 0.059697 (rank : 41) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.058806 (rank : 42) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.058530 (rank : 43) | θ value | 0.00665767 (rank : 27) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.057490 (rank : 44) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.056878 (rank : 45) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.056280 (rank : 46) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.056242 (rank : 47) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.056148 (rank : 48) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.055866 (rank : 49) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.055846 (rank : 50) | θ value | 1.06291 (rank : 80) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.055292 (rank : 51) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 130 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.055175 (rank : 52) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.055081 (rank : 53) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.055076 (rank : 54) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.054823 (rank : 55) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.054248 (rank : 56) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
COVA1_MOUSE
|
||||||
| NC score | 0.053980 (rank : 57) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.053686 (rank : 58) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.053489 (rank : 59) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.053322 (rank : 60) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.053140 (rank : 61) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.053065 (rank : 62) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.053019 (rank : 63) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.052888 (rank : 64) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.052834 (rank : 65) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.052588 (rank : 66) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.052464 (rank : 67) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.052451 (rank : 68) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.051637 (rank : 69) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.051445 (rank : 70) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.051121 (rank : 71) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.051000 (rank : 72) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.050535 (rank : 73) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.050393 (rank : 74) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.050307 (rank : 75) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MACOI_HUMAN
|
||||||
| NC score | 0.049957 (rank : 76) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.049871 (rank : 77) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.049682 (rank : 78) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.049255 (rank : 79) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.049249 (rank : 80) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.049167 (rank : 81) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.048940 (rank : 82) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.048924 (rank : 83) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.048868 (rank : 84) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.048726 (rank : 85) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.048717 (rank : 86) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.048438 (rank : 87) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.048410 (rank : 88) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.048372 (rank : 89) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.047825 (rank : 90) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.047739 (rank : 91) | θ value | 0.21417 (rank : 45) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.047530 (rank : 92) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.047423 (rank : 93) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.047321 (rank : 94) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.046983 (rank : 95) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.046287 (rank : 96) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.046193 (rank : 97) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.046184 (rank : 98) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.045732 (rank : 99) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.045384 (rank : 100) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
PCIF1_HUMAN
|
||||||
| NC score | 0.045325 (rank : 101) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
CTGE5_MOUSE
|
||||||
| NC score | 0.044898 (rank : 102) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
HOME1_HUMAN
|
||||||
| NC score | 0.044192 (rank : 103) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1. | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.042943 (rank : 104) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.042220 (rank : 105) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.041576 (rank : 106) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.041075 (rank : 107) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.040890 (rank : 108) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.040291 (rank : 109) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.040262 (rank : 110) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.039853 (rank : 111) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.038189 (rank : 112) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.038145 (rank : 113) | θ value | 1.06291 (rank : 78) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.038117 (rank : 114) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.037978 (rank : 115) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.037481 (rank : 116) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.037475 (rank : 117) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.036901 (rank : 118) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.036140 (rank : 119) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.036094 (rank : 120) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
SYWM_MOUSE
|
||||||
| NC score | 0.035285 (rank : 121) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYK1 | Gene names | Wars2 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptophanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) ((Mt)TrpRS). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.034765 (rank : 122) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.033534 (rank : 123) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.033426 (rank : 124) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.032573 (rank : 125) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.031849 (rank : 126) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.031819 (rank : 127) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.031077 (rank : 128) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.030770 (rank : 129) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
WBP4_HUMAN
|
||||||
| NC score | 0.030029 (rank : 130) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75554 | Gene names | WBP4, FBP21, FNBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.029957 (rank : 131) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.029644 (rank : 132) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.028971 (rank : 133) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.028526 (rank : 134) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CT116_HUMAN
|
||||||
| NC score | 0.027004 (rank : 135) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96HY6, Q9BW47 | Gene names | C20orf116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 precursor. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.026817 (rank : 136) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PSIP1_HUMAN
|
||||||
| NC score | 0.026448 (rank : 137) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
UT14B_MOUSE
|
||||||
| NC score | 0.025971 (rank : 138) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
FLNA_MOUSE
|
||||||
| NC score | 0.025519 (rank : 139) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.025418 (rank : 140) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
GBP2_MOUSE
|
||||||
| NC score | 0.024763 (rank : 141) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Z0E6, Q8CIC6, Q921N2, Q9R1I0 | Gene names | Gbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (mGBP2) (mGBP-2). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.024437 (rank : 142) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.024070 (rank : 143) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
K2C3_HUMAN
|
||||||
| NC score | 0.023851 (rank : 144) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.023584 (rank : 145) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.023349 (rank : 146) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.022136 (rank : 147) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.020907 (rank : 148) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
K2C4_MOUSE
|
||||||
| NC score | 0.020387 (rank : 149) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.020136 (rank : 150) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.019840 (rank : 151) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ST18_HUMAN
|
||||||
| NC score | 0.019638 (rank : 152) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
ARS2_HUMAN
|
||||||
| NC score | 0.018871 (rank : 153) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
CCD38_MOUSE
|
||||||
| NC score | 0.015926 (rank : 154) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CDN8, Q8CDD6, Q8CDE1 | Gene names | Ccdc38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.015753 (rank : 155) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
PSA7_HUMAN
|
||||||
| NC score | 0.015558 (rank : 156) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14818, Q9BR53, Q9H4K5, Q9UEU8 | Gene names | PSMA7 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 7 (EC 3.4.25.1) (Proteasome subunit RC6- 1) (Proteasome subunit XAPC7). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.015259 (rank : 157) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.015208 (rank : 158) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PSA7_MOUSE
|
||||||
| NC score | 0.014128 (rank : 159) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2U0 | Gene names | Psma7 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit alpha type 7 (EC 3.4.25.1) (Proteasome subunit RC6- 1). | |||||
|
TEX15_HUMAN
|
||||||
| NC score | 0.014099 (rank : 160) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
RWDD1_MOUSE
|
||||||
| NC score | 0.013669 (rank : 161) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQK7 | Gene names | Rwdd1 | |||
|
Domain Architecture |

|
|||||
| Description | RWD domain-containing protein 1 (IH1). | |||||
|
RWDD1_HUMAN
|
||||||
| NC score | 0.013326 (rank : 162) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H446, Q9Y313, Q9Y6B3 | Gene names | RWDD1 | |||
|
Domain Architecture |

|
|||||
| Description | RWD domain-containing protein 1. | |||||
|
TRML1_HUMAN
|
||||||
| NC score | 0.011398 (rank : 163) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YW5, Q496B3, Q8IWY1, Q8IWY2 | Gene names | TREML1, TLT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trem-like transcript 1 protein precursor (TLT-1) (Triggering receptor expressed on myeloid cells-like protein 1). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.009017 (rank : 164) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SGOL1_MOUSE
|
||||||
| NC score | 0.008734 (rank : 165) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CXH7, Q3U4K4, Q588H1, Q8BKW2 | Gene names | Sgol1, Sgo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1. | |||||
|
PABP4_HUMAN
|
||||||
| NC score | 0.008216 (rank : 166) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13310, Q6P0N3 | Gene names | PABPC4, APP1, PABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 4 (Poly(A)-binding protein 4) (PABP 4) (Inducible poly(A)-binding protein) (iPABP) (Activated-platelet protein 1) (APP-1). | |||||
|
FA40A_MOUSE
|
||||||
| NC score | 0.007371 (rank : 167) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C079, Q80T92, Q8BZC6, Q8CIF7 | Gene names | Fam40a, Kiaa1761 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM40A. | |||||
|
IRX5_MOUSE
|
||||||
| NC score | 0.006839 (rank : 168) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKQ4, Q80WV4, Q9JLL5 | Gene names | Irx5, Irxb2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2). | |||||
|
KIF1C_MOUSE
|
||||||
| NC score | 0.001869 (rank : 169) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.001191 (rank : 170) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
IKKB_MOUSE
|
||||||
| NC score | 0.000118 (rank : 171) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88351, Q9R1J6 | Gene names | Ikbkb, Ikkb | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
IKKB_HUMAN
|
||||||
| NC score | -0.000534 (rank : 172) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 166 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14920, O75327 | Gene names | IKBKB, IKKB | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||