Please be patient as the page loads
|
TEX15_HUMAN
|
||||||
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TEX15_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 174 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 2) | NC score | 0.113856 (rank : 4) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
AF9_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 3) | NC score | 0.126025 (rank : 3) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 4) | NC score | 0.071161 (rank : 12) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 5) | NC score | 0.126342 (rank : 2) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.100966 (rank : 6) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.059380 (rank : 35) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.060247 (rank : 30) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.060915 (rank : 27) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CI084_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.101737 (rank : 5) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VXU9, Q96M73 | Gene names | C9orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf84. | |||||
|
PDE1C_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.045678 (rank : 85) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
ASPM_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.066273 (rank : 19) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CJ27, O88482, Q8BJI8, Q8BKT4 | Gene names | Aspm, Calmbp1, Sha1 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein homolog (Calmodulin-binding protein 1) (Spindle and hydroxyurea checkpoint abnormal protein) (Calmodulin-binding protein Sha1). | |||||
|
ESCO1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.068170 (rank : 16) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.067784 (rank : 18) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
PLCD1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.051570 (rank : 70) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3B1 | Gene names | Plcd1, Plcd | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.024432 (rank : 135) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.067934 (rank : 17) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.060160 (rank : 31) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.053836 (rank : 55) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.064604 (rank : 24) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.086932 (rank : 8) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.066102 (rank : 21) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.065908 (rank : 22) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.088153 (rank : 7) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.064803 (rank : 23) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
ANKK1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.010753 (rank : 173) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.055487 (rank : 44) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.036381 (rank : 105) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.070392 (rank : 13) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FYB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.057001 (rank : 41) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.059683 (rank : 33) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.060850 (rank : 28) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.060120 (rank : 32) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.038296 (rank : 98) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
REST_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.055485 (rank : 45) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.070336 (rank : 14) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.085427 (rank : 9) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
BMPR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.017295 (rank : 155) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13873, Q16569 | Gene names | BMPR2, PPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type-2 precursor (EC 2.7.11.30) (Bone morphogenetic protein receptor type II) (BMP type II receptor) (BMPR-II). | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.060281 (rank : 29) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
OSBL6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.037383 (rank : 103) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BZF3, Q7Z4Q1, Q96SR1 | Gene names | OSBPL6, ORP6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.082476 (rank : 10) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
RASA2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.035144 (rank : 107) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.066123 (rank : 20) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.034264 (rank : 109) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.047984 (rank : 79) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
EPN4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.052061 (rank : 66) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |
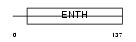
|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.051774 (rank : 67) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
RASA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.034248 (rank : 110) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
BARD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.020365 (rank : 145) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
CA103_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.053309 (rank : 60) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T3J3, Q86XS4, Q8N3B6, Q96HT4, Q9NUM5, Q9NV32 | Gene names | C1orf103, RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf103 (Receptor-interacting factor 1). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.054492 (rank : 51) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.055148 (rank : 47) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.020104 (rank : 146) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.030473 (rank : 117) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
ATF6A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.041321 (rank : 92) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.047706 (rank : 80) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.081927 (rank : 11) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.056056 (rank : 42) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ITB6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.014374 (rank : 161) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
KLC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.027555 (rank : 126) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.050601 (rank : 74) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.037384 (rank : 102) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
OSBL6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.035193 (rank : 106) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BXR9, Q8BYW2 | Gene names | Osbpl6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.053366 (rank : 59) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.062713 (rank : 25) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
ZFP37_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.003668 (rank : 187) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6Q3 | Gene names | ZFP37 | |||
|
Domain Architecture |
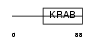
|
|||||
| Description | Zinc finger protein 37 homolog (Zfp-37). | |||||
|
ZN577_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.003519 (rank : 188) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BSK1 | Gene names | ZNF577 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 577. | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.057297 (rank : 40) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.049141 (rank : 76) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.054238 (rank : 53) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CF146_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.045944 (rank : 84) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D9W6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf146 homolog. | |||||
|
CIR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.058179 (rank : 37) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CN145_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.057908 (rank : 38) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.054550 (rank : 50) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.044316 (rank : 89) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.040819 (rank : 93) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.068311 (rank : 15) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SPB10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.011845 (rank : 170) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48595, Q4VAX4 | Gene names | SERPINB10, PI10 | |||
|
Domain Architecture |
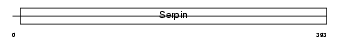
|
|||||
| Description | Serpin B10 (Bomapin) (Protease inhibitor 10). | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.033951 (rank : 111) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
ZBTB6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.009150 (rank : 176) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K088, Q8BLM4 | Gene names | Zbtb6, Zfp482, Znf482 | |||
|
Domain Architecture |
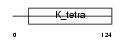
|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482). | |||||
|
BMPR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.014654 (rank : 160) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35607 | Gene names | Bmpr2 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type-2 precursor (EC 2.7.11.30) (Bone morphogenetic protein receptor type II) (BMP type II receptor) (BMPR-II) (BRK-3). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.046205 (rank : 81) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
JIP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.038564 (rank : 96) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
K22E_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.019548 (rank : 147) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
PLCD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.037782 (rank : 100) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51178 | Gene names | PLCD1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
TSH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.030819 (rank : 116) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
AATM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.023862 (rank : 136) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05202, O09188 | Gene names | Got2, Got-2 | |||
|
Domain Architecture |
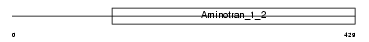
|
|||||
| Description | Aspartate aminotransferase, mitochondrial precursor (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 2). | |||||
|
BNIP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.032570 (rank : 113) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12983, O14620, Q96GP0 | Gene names | BNIP3, NIP3 | |||
|
Domain Architecture |
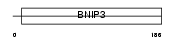
|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3. | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.024910 (rank : 132) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.059454 (rank : 34) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.046144 (rank : 82) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
EPIPL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.021823 (rank : 139) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
HCDH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.025843 (rank : 129) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |
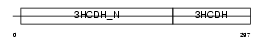
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.028076 (rank : 125) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
MASTL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.006198 (rank : 184) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C0P0, Q5RJW0, Q6NXX9, Q8BVF3, Q9CZH9, Q9D9V0 | Gene names | Mastl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.053787 (rank : 56) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.029996 (rank : 120) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.029374 (rank : 121) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
ROR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.003105 (rank : 189) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01973, Q92776 | Gene names | ROR1, NTRKR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR1 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 1). | |||||
|
RSSA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.044861 (rank : 87) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08865, P11085, P12030, Q16471, Q6IPD1 | Gene names | RPSA, LAMBR, LAMR1 | |||
|
Domain Architecture |

|
|||||
| Description | 40S ribosomal protein SA (p40) (34/67 kDa laminin receptor) (Colon carcinoma laminin-binding protein) (NEM/1CHD4) (Multidrug resistance- associated protein MGr1-Ag). | |||||
|
RSSA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.044873 (rank : 86) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14206 | Gene names | Rpsa, Lamr1, P40-8 | |||
|
Domain Architecture |

|
|||||
| Description | 40S ribosomal protein SA (p40) (34/67 kDa laminin receptor). | |||||
|
SON_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.030179 (rank : 119) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.007282 (rank : 180) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||
|
ZBTB6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.008036 (rank : 178) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |
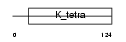
|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||
|
ZN700_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.002446 (rank : 190) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0M5 | Gene names | ZNF700 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 700. | |||||
|
CT043_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.029228 (rank : 122) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K95, Q543D3, Q80X88, Q9CV27 | Gene names | ||||
|
Domain Architecture |
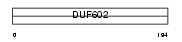
|
|||||
| Description | Uncharacterized protein C20orf43 homolog. | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.012053 (rank : 169) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
INADL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.010989 (rank : 172) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.017550 (rank : 151) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.055929 (rank : 43) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.054354 (rank : 52) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.026382 (rank : 128) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.032228 (rank : 114) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
P80C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.037498 (rank : 101) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
PALB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.031271 (rank : 115) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YC2, Q8N7Y6, Q8ND31, Q9H6W1 | Gene names | PALB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.039603 (rank : 94) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.027062 (rank : 127) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SFRP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.016916 (rank : 157) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97299, O08862, O35297 | Gene names | Sfrp2, Sarp1, Sdf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (SDF5 protein). | |||||
|
SPB10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.009575 (rank : 175) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1K6 | Gene names | Serpinb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B10. | |||||
|
TARA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.054617 (rank : 49) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.008035 (rank : 179) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TSH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.024479 (rank : 134) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.046072 (rank : 83) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.049865 (rank : 75) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
BUB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.018143 (rank : 149) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
CAPS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.016256 (rank : 158) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UW7, Q658Q2, Q7Z5T7, Q8IZW9, Q8N7M4, Q9H6P4, Q9HCI1, Q9NWK8 | Gene names | CADPS2, CAPS2, KIAA1591 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.051681 (rank : 68) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
DVL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.018723 (rank : 148) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |
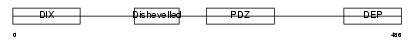
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
FKSG2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.022342 (rank : 138) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAU6 | Gene names | FKSG2 | |||
|
Domain Architecture |
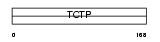
|
|||||
| Description | Apoptosis inhibitor FKSG2. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.038255 (rank : 99) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
HSP7C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.013817 (rank : 164) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.014099 (rank : 163) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
I15RA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.028260 (rank : 124) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13261, Q6B0J2, Q7LDR4, Q7Z609 | Gene names | IL15RA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-15 receptor alpha chain precursor (IL-15R-alpha) (IL- 15RA). | |||||
|
IMB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.017466 (rank : 153) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70168, Q62117 | Gene names | Kpnb1, Impnb | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Pore targeting complex 97 kDa subunit) (PTAC97) (SCG). | |||||
|
INOC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.030300 (rank : 118) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.038396 (rank : 97) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NOL8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.023551 (rank : 137) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.035133 (rank : 108) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
POLI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.017138 (rank : 156) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6R3M4, Q641P1, Q6R3M3, Q9R1A6 | Gene names | Poli, Rad30b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (Rad30 homolog B). | |||||
|
RPB6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.041967 (rank : 91) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61218, P41584 | Gene names | POLR2F, POLRF | |||
|
Domain Architecture |
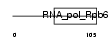
|
|||||
| Description | DNA-directed RNA polymerases I, II, and III 14.4 kDa polypeptide (EC 2.7.7.6) (RPB6) (RPABC14.4) (RPB14.4) (RPABC2). | |||||
|
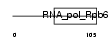
RPB6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.041981 (rank : 90) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61219 | Gene names | Polr2f | |||
|
Domain Architecture |
|
|||||
| Description | DNA-directed RNA polymerases I, II, and III 14.4 kDa polypeptide (EC 2.7.7.6) (RPB6) (RPABC14.4) (RPB14.4). | |||||
|
SFRP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.015589 (rank : 159) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HF1, O14778, Q9HAP5 | Gene names | SFRP2, FRP2, SARP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (Frizzled-related protein 2) (FRP-2). | |||||
|
SGOL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.049070 (rank : 77) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
SH24A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.033419 (rank : 112) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
SP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.006341 (rank : 183) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70494, Q68FF2, Q8CF64, Q8K378 | Gene names | Sp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp3. | |||||
|
STC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.020707 (rank : 144) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O76061 | Gene names | STC2 | |||
|
Domain Architecture |

|
|||||
| Description | Stanniocalcin-2 precursor (STC-2) (Stanniocalcin-related protein) (STCRP) (STC-related protein). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.013534 (rank : 165) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
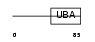
UBP2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.025157 (rank : 131) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.037342 (rank : 104) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ABLM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.005116 (rank : 185) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
ANC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.021525 (rank : 141) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H1A4, Q9BSE6, Q9H8D0 | Gene names | ANAPC1, TSG24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
ANC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.021697 (rank : 140) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.020845 (rank : 143) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ASPM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.044413 (rank : 88) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
BTBD7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.013205 (rank : 167) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
CA026_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.025316 (rank : 130) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.017536 (rank : 152) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.029081 (rank : 123) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
DVL1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.017620 (rank : 150) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
DVL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.017324 (rank : 154) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
FOXF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.007204 (rank : 182) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12946, Q5FWE5 | Gene names | FOXF1, FKHL5, FREAC1 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein F1 (Forkhead-related protein FKHL5) (Forkhead- related transcription factor 1) (FREAC-1) (Forkhead-related activator 1). | |||||
|
FOXF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.007241 (rank : 181) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61080, Q61661 | Gene names | Foxf1, Fkhl5, Foxf1a, Freac1, Hfh8 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein F1 (Forkhead-related protein FKHL5) (Forkhead- related transcription factor 1) (FREAC-1) (Hepatocyte nuclear factor 3 forkhead homolog 8) (HFH-8). | |||||
|
FUS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.013348 (rank : 166) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
GSTM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.008337 (rank : 177) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15626 | Gene names | Gstm2 | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione S-transferase Mu 2 (EC 2.5.1.18) (GST class-mu 2) (Glutathione S-transferase pmGT2) (GST 5-5). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.013073 (rank : 168) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.038658 (rank : 95) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NPT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.011079 (rank : 171) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14916, Q13783 | Gene names | SLC17A1, NPT1 | |||
|
Domain Architecture |
|
|||||
| Description | Sodium-dependent phosphate transport protein 1 (Sodium/phosphate cotransporter 1) (Na(+)/PI cotransporter 1) (Solute carrier family 17 member 1) (Renal sodium-dependent phosphate transport protein 1) (Renal sodium-phosphate transport protein 1) (Renal Na(+)-dependent phosphate cotransporter 1) (Na/Pi-4). | |||||
|
RAB3I_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.024762 (rank : 133) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
RNS10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.021196 (rank : 142) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D5A9, Q6XL64 | Gene names | Rnase10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease-like protein 10 precursor (Protein Train A). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.048265 (rank : 78) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.014249 (rank : 162) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.052991 (rank : 61) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.010317 (rank : 174) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
VLDLR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.003962 (rank : 186) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.051031 (rank : 72) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CF060_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.050937 (rank : 73) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.052322 (rank : 64) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.058918 (rank : 36) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.053623 (rank : 57) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.051124 (rank : 71) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.054936 (rank : 48) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
MAP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.053419 (rank : 58) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.057667 (rank : 39) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.054185 (rank : 54) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.062263 (rank : 26) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
REST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.051677 (rank : 69) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.052698 (rank : 63) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.052910 (rank : 62) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.052220 (rank : 65) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.055389 (rank : 46) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
TEX15_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 174 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.126342 (rank : 2) | θ value | 0.0193708 (rank : 5) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.126025 (rank : 3) | θ value | 0.00390308 (rank : 3) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.113856 (rank : 4) | θ value | 0.000461057 (rank : 2) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CI084_HUMAN
|
||||||
| NC score | 0.101737 (rank : 5) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VXU9, Q96M73 | Gene names | C9orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf84. | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.100966 (rank : 6) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.088153 (rank : 7) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.086932 (rank : 8) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.085427 (rank : 9) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.082476 (rank : 10) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.081927 (rank : 11) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.071161 (rank : 12) | θ value | 0.0148317 (rank : 4) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.070392 (rank : 13) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.070336 (rank : 14) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.068311 (rank : 15) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
ESCO1_MOUSE
|
||||||
| NC score | 0.068170 (rank : 16) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.067934 (rank : 17) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.067784 (rank : 18) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ASPM_MOUSE
|
||||||
| NC score | 0.066273 (rank : 19) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CJ27, O88482, Q8BJI8, Q8BKT4 | Gene names | Aspm, Calmbp1, Sha1 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein homolog (Calmodulin-binding protein 1) (Spindle and hydroxyurea checkpoint abnormal protein) (Calmodulin-binding protein Sha1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.066123 (rank : 20) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.066102 (rank : 21) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.065908 (rank : 22) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.064803 (rank : 23) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.064604 (rank : 24) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.062713 (rank : 25) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.062263 (rank : 26) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.060915 (rank : 27) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.060850 (rank : 28) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.060281 (rank : 29) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.060247 (rank : 30) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.060160 (rank : 31) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.060120 (rank : 32) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.059683 (rank : 33) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.059454 (rank : 34) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.059380 (rank : 35) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.058918 (rank : 36) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.058179 (rank : 37) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.057908 (rank : 38) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.057667 (rank : 39) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.057297 (rank : 40) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.057001 (rank : 41) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.056056 (rank : 42) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.055929 (rank : 43) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.055487 (rank : 44) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.055485 (rank : 45) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.055389 (rank : 46) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.055148 (rank : 47) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.054936 (rank : 48) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.054617 (rank : 49) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.054550 (rank : 50) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.054492 (rank : 51) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.054354 (rank : 52) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.054238 (rank : 53) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.054185 (rank : 54) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.053836 (rank : 55) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.053787 (rank : 56) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.053623 (rank : 57) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.053419 (rank : 58) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.053366 (rank : 59) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
CA103_HUMAN
|
||||||
| NC score | 0.053309 (rank : 60) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T3J3, Q86XS4, Q8N3B6, Q96HT4, Q9NUM5, Q9NV32 | Gene names | C1orf103, RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf103 (Receptor-interacting factor 1). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.052991 (rank : 61) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.052910 (rank : 62) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.052698 (rank : 63) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.052322 (rank : 64) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.052220 (rank : 65) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
EPN4_HUMAN
|
||||||
| NC score | 0.052061 (rank : 66) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| NC score | 0.051774 (rank : 67) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.051681 (rank : 68) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.051677 (rank : 69) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
PLCD1_MOUSE
|
||||||
| NC score | 0.051570 (rank : 70) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3B1 | Gene names | Plcd1, Plcd | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.051124 (rank : 71) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.051031 (rank : 72) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.050937 (rank : 73) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.050601 (rank : 74) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.049865 (rank : 75) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.049141 (rank : 76) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
SGOL2_HUMAN
|
||||||
| NC score | 0.049070 (rank : 77) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.048265 (rank : 78) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.047984 (rank : 79) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.047706 (rank : 80) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.046205 (rank : 81) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.046144 (rank : 82) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.046072 (rank : 83) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
CF146_MOUSE
|
||||||
| NC score | 0.045944 (rank : 84) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D9W6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf146 homolog. | |||||
|
PDE1C_HUMAN
|
||||||
| NC score | 0.045678 (rank : 85) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14123, Q14124 | Gene names | PDE1C | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C) (hCam-3). | |||||
|
RSSA_MOUSE
|
||||||
| NC score | 0.044873 (rank : 86) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14206 | Gene names | Rpsa, Lamr1, P40-8 | |||
|
Domain Architecture |

|
|||||
| Description | 40S ribosomal protein SA (p40) (34/67 kDa laminin receptor). | |||||
|
RSSA_HUMAN
|
||||||
| NC score | 0.044861 (rank : 87) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08865, P11085, P12030, Q16471, Q6IPD1 | Gene names | RPSA, LAMBR, LAMR1 | |||
|
Domain Architecture |

|
|||||
| Description | 40S ribosomal protein SA (p40) (34/67 kDa laminin receptor) (Colon carcinoma laminin-binding protein) (NEM/1CHD4) (Multidrug resistance- associated protein MGr1-Ag). | |||||
|
ASPM_HUMAN
|
||||||
| NC score | 0.044413 (rank : 88) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.044316 (rank : 89) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
RPB6_MOUSE
|
||||||
| NC score | 0.041981 (rank : 90) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61219 | Gene names | Polr2f | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerases I, II, and III 14.4 kDa polypeptide (EC 2.7.7.6) (RPB6) (RPABC14.4) (RPB14.4). | |||||
|
RPB6_HUMAN
|
||||||
| NC score | 0.041967 (rank : 91) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61218, P41584 | Gene names | POLR2F, POLRF | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerases I, II, and III 14.4 kDa polypeptide (EC 2.7.7.6) (RPB6) (RPABC14.4) (RPB14.4) (RPABC2). | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.041321 (rank : 92) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.040819 (rank : 93) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.039603 (rank : 94) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.038658 (rank : 95) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
JIP4_MOUSE
|
||||||
| NC score | 0.038564 (rank : 96) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.038396 (rank : 97) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.038296 (rank : 98) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.038255 (rank : 99) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
PLCD1_HUMAN
|
||||||
| NC score | 0.037782 (rank : 100) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51178 | Gene names | PLCD1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.037498 (rank : 101) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.037384 (rank : 102) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
OSBL6_HUMAN
|
||||||
| NC score | 0.037383 (rank : 103) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BZF3, Q7Z4Q1, Q96SR1 | Gene names | OSBPL6, ORP6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.037342 (rank : 104) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.036381 (rank : 105) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
OSBL6_MOUSE
|
||||||
| NC score | 0.035193 (rank : 106) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BXR9, Q8BYW2 | Gene names | Osbpl6 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 6 (OSBP-related protein 6) (ORP-6). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.035144 (rank : 107) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.035133 (rank : 108) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.034264 (rank : 109) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
RASA2_HUMAN
|
||||||
| NC score | 0.034248 (rank : 110) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.033951 (rank : 111) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
SH24A_MOUSE
|
||||||
| NC score | 0.033419 (rank : 112) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
BNIP3_HUMAN
|
||||||
| NC score | 0.032570 (rank : 113) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12983, O14620, Q96GP0 | Gene names | BNIP3, NIP3 | |||
|
Domain Architecture |

|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3. | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.032228 (rank : 114) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PALB2_HUMAN
|
||||||
| NC score | 0.031271 (rank : 115) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YC2, Q8N7Y6, Q8ND31, Q9H6W1 | Gene names | PALB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
TSH2_HUMAN
|
||||||
| NC score | 0.030819 (rank : 116) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NRE2, Q4VXM4, Q6N003, Q8N260 | Gene names | TSHZ2, C20orf17, TSH2, ZNF218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (Ovarian cancer-related protein 10-2) (OVC10-2). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.030473 (rank : 117) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
INOC1_HUMAN
|
||||||
| NC score | 0.030300 (rank : 118) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.030179 (rank : 119) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.029996 (rank : 120) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.029374 (rank : 121) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
CT043_MOUSE
|
||||||
| NC score | 0.029228 (rank : 122) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K95, Q543D3, Q80X88, Q9CV27 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf43 homolog. | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.029081 (rank : 123) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
I15RA_HUMAN
|
||||||
| NC score | 0.028260 (rank : 124) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13261, Q6B0J2, Q7LDR4, Q7Z609 | Gene names | IL15RA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-15 receptor alpha chain precursor (IL-15R-alpha) (IL- 15RA). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.028076 (rank : 125) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
KLC3_HUMAN
|
||||||
| NC score | 0.027555 (rank : 126) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P597, Q6GMU2, Q8NAL1, Q8WWJ9 | Gene names | KLC3, KLC2, KLC2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin light chain 3 (kinesin light chain 2) (KLC2-like). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.027062 (rank : 127) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.026382 (rank : 128) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
HCDH_MOUSE
|
||||||
| NC score | 0.025843 (rank : 129) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
CA026_MOUSE
|
||||||
| NC score | 0.025316 (rank : 130) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DBQ9, Q6PAN2, Q7TMQ6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf26 homolog. | |||||
|
UBP2L_HUMAN
|
||||||
| NC score | 0.025157 (rank : 131) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.024910 (rank : 132) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
RAB3I_HUMAN
|
||||||
| NC score | 0.024762 (rank : 133) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
TSH2_MOUSE
|
||||||
| NC score | 0.024479 (rank : 134) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.024432 (rank : 135) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
AATM_MOUSE
|
||||||
| NC score | 0.023862 (rank : 136) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05202, O09188 | Gene names | Got2, Got-2 | |||
|
Domain Architecture |

|
|||||
| Description | Aspartate aminotransferase, mitochondrial precursor (EC 2.6.1.1) (Transaminase A) (Glutamate oxaloacetate transaminase 2). | |||||
|
NOL8_MOUSE
|
||||||
| NC score | 0.023551 (rank : 137) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
FKSG2_HUMAN
|
||||||
| NC score | 0.022342 (rank : 138) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAU6 | Gene names | FKSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis inhibitor FKSG2. | |||||
|
EPIPL_HUMAN
|
||||||
| NC score | 0.021823 (rank : 139) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
ANC1_MOUSE
|
||||||
| NC score | 0.021697 (rank : 140) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
ANC1_HUMAN
|
||||||
| NC score | 0.021525 (rank : 141) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H1A4, Q9BSE6, Q9H8D0 | Gene names | ANAPC1, TSG24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
RNS10_MOUSE
|
||||||
| NC score | 0.021196 (rank : 142) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D5A9, Q6XL64 | Gene names | Rnase10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease-like protein 10 precursor (Protein Train A). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.020845 (rank : 143) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
STC2_HUMAN
|
||||||
| NC score | 0.020707 (rank : 144) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O76061 | Gene names | STC2 | |||
|
Domain Architecture |

|
|||||
| Description | Stanniocalcin-2 precursor (STC-2) (Stanniocalcin-related protein) (STCRP) (STC-related protein). | |||||
|
BARD1_MOUSE
|
||||||
| NC score | 0.020365 (rank : 145) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | 0.020104 (rank : 146) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
K22E_HUMAN
|
||||||
| NC score | 0.019548 (rank : 147) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 515 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35908 | Gene names | KRT2A, KRT2E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 epidermal (Cytokeratin-2e) (K2e) (CK 2e). | |||||
|
DVL1_MOUSE
|
||||||
| NC score | 0.018723 (rank : 148) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
BUB1_HUMAN
|
||||||
| NC score | 0.018143 (rank : 149) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
DVL1L_HUMAN
|
||||||
| NC score | 0.017620 (rank : 150) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.017550 (rank : 151) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.017536 (rank : 152) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
IMB1_MOUSE
|
||||||
| NC score | 0.017466 (rank : 153) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70168, Q62117 | Gene names | Kpnb1, Impnb | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Pore targeting complex 97 kDa subunit) (PTAC97) (SCG). | |||||
|
DVL1_HUMAN
|
||||||
| NC score | 0.017324 (rank : 154) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
BMPR2_HUMAN
|
||||||
| NC score | 0.017295 (rank : 155) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13873, Q16569 | Gene names | BMPR2, PPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type-2 precursor (EC 2.7.11.30) (Bone morphogenetic protein receptor type II) (BMP type II receptor) (BMPR-II). | |||||
|
POLI_MOUSE
|
||||||
| NC score | 0.017138 (rank : 156) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6R3M4, Q641P1, Q6R3M3, Q9R1A6 | Gene names | Poli, Rad30b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (Rad30 homolog B). | |||||
|
SFRP2_MOUSE
|
||||||
| NC score | 0.016916 (rank : 157) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97299, O08862, O35297 | Gene names | Sfrp2, Sarp1, Sdf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (SDF5 protein). | |||||
|
CAPS2_HUMAN
|
||||||
| NC score | 0.016256 (rank : 158) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UW7, Q658Q2, Q7Z5T7, Q8IZW9, Q8N7M4, Q9H6P4, Q9HCI1, Q9NWK8 | Gene names | CADPS2, CAPS2, KIAA1591 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
SFRP2_HUMAN
|
||||||
| NC score | 0.015589 (rank : 159) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HF1, O14778, Q9HAP5 | Gene names | SFRP2, FRP2, SARP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (Frizzled-related protein 2) (FRP-2). | |||||
|
BMPR2_MOUSE
|
||||||
| NC score | 0.014654 (rank : 160) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35607 | Gene names | Bmpr2 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type-2 precursor (EC 2.7.11.30) (Bone morphogenetic protein receptor type II) (BMP type II receptor) (BMPR-II) (BRK-3). | |||||
|
ITB6_HUMAN
|
||||||
| NC score | 0.014374 (rank : 161) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.014249 (rank : 162) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
HSP7C_MOUSE
|
||||||
| NC score | 0.014099 (rank : 163) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_HUMAN
|
||||||
| NC score | 0.013817 (rank : 164) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.013534 (rank : 165) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.013348 (rank : 166) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
BTBD7_MOUSE
|
||||||
| NC score | 0.013205 (rank : 167) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.013073 (rank : 168) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.012053 (rank : 169) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
SPB10_HUMAN
|
||||||
| NC score | 0.011845 (rank : 170) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48595, Q4VAX4 | Gene names | SERPINB10, PI10 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B10 (Bomapin) (Protease inhibitor 10). | |||||
|
NPT1_HUMAN
|
||||||
| NC score | 0.011079 (rank : 171) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14916, Q13783 | Gene names | SLC17A1, NPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent phosphate transport protein 1 (Sodium/phosphate cotransporter 1) (Na(+)/PI cotransporter 1) (Solute carrier family 17 member 1) (Renal sodium-dependent phosphate transport protein 1) (Renal sodium-phosphate transport protein 1) (Renal Na(+)-dependent phosphate cotransporter 1) (Na/Pi-4). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.010989 (rank : 172) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
ANKK1_HUMAN
|
||||||
| NC score | 0.010753 (rank : 173) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1075 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NFD2 | Gene names | ANKK1, PKK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and protein kinase domain-containing protein 1 (EC 2.7.11.1) (Protein kinase PKK2) (X-kinase) (SgK288). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.010317 (rank : 174) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
SPB10_MOUSE
|
||||||
| NC score | 0.009575 (rank : 175) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1K6 | Gene names | Serpinb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin B10. | |||||
|
ZBTB6_MOUSE
|
||||||
| NC score | 0.009150 (rank : 176) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K088, Q8BLM4 | Gene names | Zbtb6, Zfp482, Znf482 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482). | |||||
|
GSTM2_MOUSE
|
||||||
| NC score | 0.008337 (rank : 177) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15626 | Gene names | Gstm2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase Mu 2 (EC 2.5.1.18) (GST class-mu 2) (Glutathione S-transferase pmGT2) (GST 5-5). | |||||
|
ZBTB6_HUMAN
|
||||||
| NC score | 0.008036 (rank : 178) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.008035 (rank : 179) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
SP3_HUMAN
|
||||||
| NC score | 0.007282 (rank : 180) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||
|
FOXF1_MOUSE
|
||||||
| NC score | 0.007241 (rank : 181) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61080, Q61661 | Gene names | Foxf1, Fkhl5, Foxf1a, Freac1, Hfh8 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein F1 (Forkhead-related protein FKHL5) (Forkhead- related transcription factor 1) (FREAC-1) (Hepatocyte nuclear factor 3 forkhead homolog 8) (HFH-8). | |||||
|
FOXF1_HUMAN
|
||||||
| NC score | 0.007204 (rank : 182) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12946, Q5FWE5 | Gene names | FOXF1, FKHL5, FREAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein F1 (Forkhead-related protein FKHL5) (Forkhead- related transcription factor 1) (FREAC-1) (Forkhead-related activator 1). | |||||
|
SP3_MOUSE
|
||||||
| NC score | 0.006341 (rank : 183) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70494, Q68FF2, Q8CF64, Q8K378 | Gene names | Sp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp3. | |||||
|
MASTL_MOUSE
|
||||||
| NC score | 0.006198 (rank : 184) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C0P0, Q5RJW0, Q6NXX9, Q8BVF3, Q9CZH9, Q9D9V0 | Gene names | Mastl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
ABLM1_HUMAN
|
||||||
| NC score | 0.005116 (rank : 185) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
VLDLR_HUMAN
|
||||||
| NC score | 0.003962 (rank : 186) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
ZFP37_HUMAN
|
||||||
| NC score | 0.003668 (rank : 187) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6Q3 | Gene names | ZFP37 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 37 homolog (Zfp-37). | |||||
|
ZN577_HUMAN
|
||||||
| NC score | 0.003519 (rank : 188) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BSK1 | Gene names | ZNF577 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 577. | |||||
|
ROR1_HUMAN
|
||||||
| NC score | 0.003105 (rank : 189) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01973, Q92776 | Gene names | ROR1, NTRKR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR1 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 1). | |||||
|
ZN700_HUMAN
|
||||||
| NC score | 0.002446 (rank : 190) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0M5 | Gene names | ZNF700 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 700. | |||||