Please be patient as the page loads
|
EPN4_HUMAN
|
||||||
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |
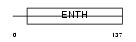
|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EPN4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992100 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 6.15952e-41 (rank : 3) | NC score | 0.809103 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN2_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 4) | NC score | 0.824570 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 1.16164e-39 (rank : 5) | NC score | 0.838251 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 2.58786e-39 (rank : 6) | NC score | 0.779611 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 7) | NC score | 0.795415 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 2.50655e-34 (rank : 8) | NC score | 0.796798 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
RU17_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.062635 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
ITPR1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.048904 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
AP180_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.117761 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
AP180_MOUSE
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.131697 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.063029 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.047994 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.026291 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
RU17_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.057370 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.059437 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PICAL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.090463 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |
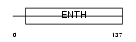
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
PICA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.090917 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7M6Y3, Q3TS04, Q811P1, Q8BUF6, Q8CIH8, Q8R0A9, Q8R3E1, Q8VDN5, Q921L0 | Gene names | Picalm, Calm, Fit1 | |||
|
Domain Architecture |
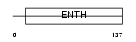
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia) (CALM). | |||||
|
RPO3G_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.063898 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15318 | Gene names | POLR3G, RPC32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.021237 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.039611 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
GAK_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.007788 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
TEX15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.052061 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.043224 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.038757 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.051255 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.035915 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
TOM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.019026 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.040120 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.035063 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.049008 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
AN32B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.023798 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.016013 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.040958 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.021866 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.020042 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
CN037_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.018663 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
DIP2C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.017541 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2E4 | Gene names | DIP2C, KIAA0934 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog C. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.014847 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
CADH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.001928 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.031718 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
FOXF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.004460 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12947, Q9UQ85 | Gene names | FOXF2, FKHL6, FREAC2 | |||
|
Domain Architecture |
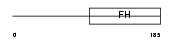
|
|||||
| Description | Forkhead box protein F2 (Forkhead-related protein FKHL6) (Forkhead- related transcription factor 2) (FREAC-2) (Forkhead-related activator 2). | |||||
|
K0494_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.010509 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGQ6, Q3TQN6, Q8BJV7, Q8C0R1 | Gene names | Kiaa0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
MEF2D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.024511 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |
|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
RPO3G_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.043998 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NXY9, Q8K0W5, Q9CV05 | Gene names | Polr3g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.017455 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.014143 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.013889 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.018188 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.071264 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.055720 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.053399 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
EPN4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| NC score | 0.992100 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.838251 (rank : 3) | θ value | 1.16164e-39 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
EPN2_HUMAN
|
||||||
| NC score | 0.824570 (rank : 4) | θ value | 8.89434e-40 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.809103 (rank : 5) | θ value | 6.15952e-41 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.796798 (rank : 6) | θ value | 2.50655e-34 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.795415 (rank : 7) | θ value | 2.50655e-34 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.779611 (rank : 8) | θ value | 2.58786e-39 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
AP180_MOUSE
|
||||||
| NC score | 0.131697 (rank : 9) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.117761 (rank : 10) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
PICA_MOUSE
|
||||||
| NC score | 0.090917 (rank : 11) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7M6Y3, Q3TS04, Q811P1, Q8BUF6, Q8CIH8, Q8R0A9, Q8R3E1, Q8VDN5, Q921L0 | Gene names | Picalm, Calm, Fit1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia) (CALM). | |||||
|
PICAL_HUMAN
|
||||||
| NC score | 0.090463 (rank : 12) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.071264 (rank : 13) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
RPO3G_HUMAN
|
||||||
| NC score | 0.063898 (rank : 14) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15318 | Gene names | POLR3G, RPC32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.063029 (rank : 15) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
RU17_HUMAN
|
||||||
| NC score | 0.062635 (rank : 16) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.059437 (rank : 17) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RU17_MOUSE
|
||||||
| NC score | 0.057370 (rank : 18) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.055720 (rank : 19) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.053399 (rank : 20) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
TEX15_HUMAN
|
||||||
| NC score | 0.052061 (rank : 21) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.051255 (rank : 22) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.049008 (rank : 23) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ITPR1_HUMAN
|
||||||
| NC score | 0.048904 (rank : 24) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.047994 (rank : 25) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
RPO3G_MOUSE
|
||||||
| NC score | 0.043998 (rank : 26) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NXY9, Q8K0W5, Q9CV05 | Gene names | Polr3g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase III subunit G (EC 2.7.7.6) (DNA-directed RNA polymerase III 32 kDa polypeptide) (RNA polymerase III C32 subunit). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.043224 (rank : 27) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.040958 (rank : 28) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.040120 (rank : 29) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.039611 (rank : 30) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.038757 (rank : 31) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.035915 (rank : 32) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.035063 (rank : 33) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.031718 (rank : 34) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.026291 (rank : 35) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
MEF2D_HUMAN
|
||||||
| NC score | 0.024511 (rank : 36) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14814, Q14815 | Gene names | MEF2D | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2D. | |||||
|
AN32B_HUMAN
|
||||||
| NC score | 0.023798 (rank : 37) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92688, O00655, P78458, P78459 | Gene names | ANP32B, APRIL, PHAPI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (PHAPI2 protein) (Silver-stainable protein SSP29) (Acidic protein rich in leucines). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.021866 (rank : 38) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.021237 (rank : 39) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.020042 (rank : 40) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
TOM1_HUMAN
|
||||||
| NC score | 0.019026 (rank : 41) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
CN037_HUMAN
|
||||||
| NC score | 0.018663 (rank : 42) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86TY3, Q6P5Q1, Q86TY1 | Gene names | C14orf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 precursor. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.018188 (rank : 43) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
DIP2C_HUMAN
|
||||||
| NC score | 0.017541 (rank : 44) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2E4 | Gene names | DIP2C, KIAA0934 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog C. | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.017455 (rank : 45) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.016013 (rank : 46) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.014847 (rank : 47) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.014143 (rank : 48) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.013889 (rank : 49) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
K0494_MOUSE
|
||||||
| NC score | 0.010509 (rank : 50) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGQ6, Q3TQN6, Q8BJV7, Q8C0R1 | Gene names | Kiaa0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.007788 (rank : 51) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
FOXF2_HUMAN
|
||||||
| NC score | 0.004460 (rank : 52) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12947, Q9UQ85 | Gene names | FOXF2, FKHL6, FREAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein F2 (Forkhead-related protein FKHL6) (Forkhead- related transcription factor 2) (FREAC-2) (Forkhead-related activator 2). | |||||
|
CADH2_MOUSE
|
||||||
| NC score | 0.001928 (rank : 53) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||