Please be patient as the page loads
|
PICA_MOUSE
|
||||||
| SwissProt Accessions | Q7M6Y3, Q3TS04, Q811P1, Q8BUF6, Q8CIH8, Q8R0A9, Q8R3E1, Q8VDN5, Q921L0 | Gene names | Picalm, Calm, Fit1 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia) (CALM). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PICAL_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994596 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
PICA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7M6Y3, Q3TS04, Q811P1, Q8BUF6, Q8CIH8, Q8R0A9, Q8R3E1, Q8VDN5, Q921L0 | Gene names | Picalm, Calm, Fit1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia) (CALM). | |||||
|
AP180_HUMAN
|
||||||
| θ value | 5.05951e-144 (rank : 3) | NC score | 0.885777 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
AP180_MOUSE
|
||||||
| θ value | 2.511e-143 (rank : 4) | NC score | 0.881107 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 5) | NC score | 0.036295 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.053851 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.038959 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.038517 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.035549 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.048693 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.054975 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.040420 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
EPN4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.090917 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
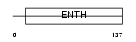
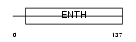
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |
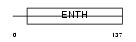
|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.090437 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
KIF22_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.019799 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14807, O94814, Q9BT46 | Gene names | KIF22, KID, KNSL4 | |||
|
Domain Architecture |
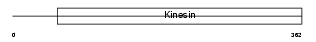
|
|||||
| Description | Kinesin-like protein KIF22 (Kinesin-like DNA-binding protein) (Kinesin-like protein 4). | |||||
|
LEG4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.036559 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K419, O88353, Q91X74 | Gene names | Lgals4 | |||
|
Domain Architecture |
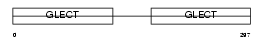
|
|||||
| Description | Galectin-4 (Lactose-binding lectin 4). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.030524 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.047575 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
DC1L2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.036945 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.059150 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.029633 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.005629 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
YTHD2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.023419 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5A9, Q5VSZ9, Q8TDH0, Q9BUJ5 | Gene names | YTHDF2, HGRG8 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 2 (High-glucose-regulated protein 8) (NY- REN-2 antigen) (CLL-associated antigen KW-14). | |||||
|
CDX1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.020233 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |
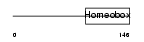
|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.022994 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.028995 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.021468 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.036601 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.029342 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.025722 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
H14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.025980 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.018008 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CASL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.020224 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |
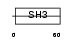
|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.003984 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.011506 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.016344 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
T2EA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.032332 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29083, Q16103 | Gene names | GTF2E1, TF2E1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE 56 kDa subunit). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.018907 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.020485 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.028695 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.014798 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.019586 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.027936 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
STK23_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.005972 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0G2 | Gene names | Stk23, Mssk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 23 (EC 2.7.11.1) (Muscle-specific serine kinase 1) (MSSK-1). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.003522 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CN032_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.024392 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.022868 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.006856 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.012047 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.013258 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
AINX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.008201 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46660, Q61958, Q8VCW5 | Gene names | Ina | |||
|
Domain Architecture |
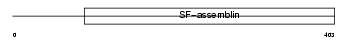
|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
CE110_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.020123 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.008565 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
EF1D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.020878 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57776, Q9CWW2, Q9CWY1, Q9CYD4, Q9CYJ5 | Gene names | Eef1d | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-delta (EF-1-delta). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.019629 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
KAD7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.016573 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2H2, Q8BVH3 | Gene names | Ak7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.016994 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.022882 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
RPP25_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.015409 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BUL9, Q9NX88 | Gene names | RPP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease P protein subunit p25 (EC 3.1.26.5) (RNase P protein subunit p25). | |||||
|
RU1C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.063185 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
RU1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.068989 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.014126 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.010284 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.059268 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.057045 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
LEUK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.062396 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.062649 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PICA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7M6Y3, Q3TS04, Q811P1, Q8BUF6, Q8CIH8, Q8R0A9, Q8R3E1, Q8VDN5, Q921L0 | Gene names | Picalm, Calm, Fit1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia) (CALM). | |||||
|
PICAL_HUMAN
|
||||||
| NC score | 0.994596 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.885777 (rank : 3) | θ value | 5.05951e-144 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
AP180_MOUSE
|
||||||
| NC score | 0.881107 (rank : 4) | θ value | 2.511e-143 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61548, Q61547 | Gene names | Snap91 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein) (Phosphoprotein F1- 20). | |||||
|
EPN4_HUMAN
|
||||||
| NC score | 0.090917 (rank : 5) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14677, Q8NAF1, Q96E05 | Gene names | CLINT1, ENTH, EPN4, EPNR, KIAA0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin) (Clathrin-interacting protein localized in the trans- Golgi region) (Clint). | |||||
|
EPN4_MOUSE
|
||||||
| NC score | 0.090437 (rank : 6) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KN9, Q8CFH4 | Gene names | Clint1, Enth, Epn4, Epnr, Kiaa0171 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin interactor 1 (Epsin-4) (Epsin-related protein) (EpsinR) (Enthoprotin). | |||||
|
RU1C_MOUSE
|
||||||
| NC score | 0.068989 (rank : 7) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
RU1C_HUMAN
|
||||||
| NC score | 0.063185 (rank : 8) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.062649 (rank : 9) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.062396 (rank : 10) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.059268 (rank : 11) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.059150 (rank : 12) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.057045 (rank : 13) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.054975 (rank : 14) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.053851 (rank : 15) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.048693 (rank : 16) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.047575 (rank : 17) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.040420 (rank : 18) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.038959 (rank : 19) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.038517 (rank : 20) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
DC1L2_MOUSE
|
||||||
| NC score | 0.036945 (rank : 21) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PDL0 | Gene names | Dync1li2, Dncli2, Dnclic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 2 (Dynein light intermediate chain 2, cytosolic). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.036601 (rank : 22) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
LEG4_MOUSE
|
||||||
| NC score | 0.036559 (rank : 23) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K419, O88353, Q91X74 | Gene names | Lgals4 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-4 (Lactose-binding lectin 4). | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.036295 (rank : 24) | θ value | 0.0431538 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.035549 (rank : 25) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
T2EA_HUMAN
|
||||||
| NC score | 0.032332 (rank : 26) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29083, Q16103 | Gene names | GTF2E1, TF2E1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE 56 kDa subunit). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.030524 (rank : 27) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.029633 (rank : 28) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.029342 (rank : 29) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.028995 (rank : 30) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.028695 (rank : 31) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.027936 (rank : 32) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.025980 (rank : 33) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.025722 (rank : 34) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.024392 (rank : 35) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
YTHD2_HUMAN
|
||||||
| NC score | 0.023419 (rank : 36) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5A9, Q5VSZ9, Q8TDH0, Q9BUJ5 | Gene names | YTHDF2, HGRG8 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 2 (High-glucose-regulated protein 8) (NY- REN-2 antigen) (CLL-associated antigen KW-14). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.022994 (rank : 37) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.022882 (rank : 38) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.022868 (rank : 39) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.021468 (rank : 40) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
EF1D_MOUSE
|
||||||
| NC score | 0.020878 (rank : 41) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57776, Q9CWW2, Q9CWY1, Q9CYD4, Q9CYJ5 | Gene names | Eef1d | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-delta (EF-1-delta). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.020485 (rank : 42) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CDX1_HUMAN
|
||||||
| NC score | 0.020233 (rank : 43) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
CASL_MOUSE
|
||||||
| NC score | 0.020224 (rank : 44) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35177, Q8BJL8, Q8BK90, Q8BL52, Q8BM94, Q8BMI9, Q99KE7 | Gene names | Nedd9, Casl | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (MEF1) (CRK-associated substrate-related protein) (CAS-L) (p105) (Protein NEDD9) (Neural precursor cell expressed developmentally down-regulated protein 9). | |||||
|
CE110_HUMAN
|
||||||
| NC score | 0.020123 (rank : 45) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43303, O43335, Q68DV9, Q8NE13 | Gene names | CEP110, CP110, KIAA0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
KIF22_HUMAN
|
||||||
| NC score | 0.019799 (rank : 46) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14807, O94814, Q9BT46 | Gene names | KIF22, KID, KNSL4 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF22 (Kinesin-like DNA-binding protein) (Kinesin-like protein 4). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.019629 (rank : 47) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.019586 (rank : 48) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.018907 (rank : 49) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.018008 (rank : 50) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.016994 (rank : 51) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
KAD7_MOUSE
|
||||||
| NC score | 0.016573 (rank : 52) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2H2, Q8BVH3 | Gene names | Ak7 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenylate kinase 7 (EC 2.7.4.3). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.016344 (rank : 53) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RPP25_HUMAN
|
||||||
| NC score | 0.015409 (rank : 54) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BUL9, Q9NX88 | Gene names | RPP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease P protein subunit p25 (EC 3.1.26.5) (RNase P protein subunit p25). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.014798 (rank : 55) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.014126 (rank : 56) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.013258 (rank : 57) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.012047 (rank : 58) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.011506 (rank : 59) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.010284 (rank : 60) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.008565 (rank : 61) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
AINX_MOUSE
|
||||||
| NC score | 0.008201 (rank : 62) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46660, Q61958, Q8VCW5 | Gene names | Ina | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.006856 (rank : 63) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
STK23_MOUSE
|
||||||
| NC score | 0.005972 (rank : 64) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0G2 | Gene names | Stk23, Mssk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 23 (EC 2.7.11.1) (Muscle-specific serine kinase 1) (MSSK-1). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.005629 (rank : 65) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.003984 (rank : 66) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.003522 (rank : 67) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||