Please be patient as the page loads
|
T2EA_HUMAN
|
||||||
| SwissProt Accessions | P29083, Q16103 | Gene names | GTF2E1, TF2E1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE 56 kDa subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
T2EA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29083, Q16103 | Gene names | GTF2E1, TF2E1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE 56 kDa subunit). | |||||
|
T2EA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994206 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D0D5, Q544T0 | Gene names | Gtf2e1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE 56 kDa subunit). | |||||
|
ZN700_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 3) | NC score | 0.011948 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M5 | Gene names | ZNF700 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 700. | |||||
|
ROBO2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 4) | NC score | 0.019143 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
DSG2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 5) | NC score | 0.013799 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55111, Q8K069, Q8R517 | Gene names | Dsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor. | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.051107 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
GAS8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 7) | NC score | 0.017767 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
K0460_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.038164 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
ZN440_HUMAN
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.006692 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYI8, Q8N1R9 | Gene names | ZNF440 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 440. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 10) | NC score | 0.020990 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PAP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.019080 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
PICAL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.032380 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |
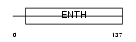
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
PICA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.032332 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M6Y3, Q3TS04, Q811P1, Q8BUF6, Q8CIH8, Q8R0A9, Q8R3E1, Q8VDN5, Q921L0 | Gene names | Picalm, Calm, Fit1 | |||
|
Domain Architecture |
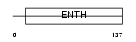
|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia) (CALM). | |||||
|
XPC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.026265 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.021193 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.020977 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.017265 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
ZN611_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.005193 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N823, Q69YG9 | Gene names | ZNF611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 611. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.014615 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
T2EA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29083, Q16103 | Gene names | GTF2E1, TF2E1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE 56 kDa subunit). | |||||
|
T2EA_MOUSE
|
||||||
| NC score | 0.994206 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D0D5, Q544T0 | Gene names | Gtf2e1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE 56 kDa subunit). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.051107 (rank : 3) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.038164 (rank : 4) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
PICAL_HUMAN
|
||||||
| NC score | 0.032380 (rank : 5) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13492, O60700, Q86XZ9 | Gene names | PICALM, CALM | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia protein). | |||||
|
PICA_MOUSE
|
||||||
| NC score | 0.032332 (rank : 6) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M6Y3, Q3TS04, Q811P1, Q8BUF6, Q8CIH8, Q8R0A9, Q8R3E1, Q8VDN5, Q921L0 | Gene names | Picalm, Calm, Fit1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-binding clathrin assembly protein (Clathrin assembly lymphoid myeloid leukemia) (CALM). | |||||
|
XPC_MOUSE
|
||||||
| NC score | 0.026265 (rank : 7) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.021193 (rank : 8) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.020990 (rank : 9) | θ value | 4.03905 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.020977 (rank : 10) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
ROBO2_MOUSE
|
||||||
| NC score | 0.019143 (rank : 11) | θ value | 0.0961366 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
PAP2_HUMAN
|
||||||
| NC score | 0.019080 (rank : 12) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
GAS8_MOUSE
|
||||||
| NC score | 0.017767 (rank : 13) | θ value | 3.0926 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.017265 (rank : 14) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.014615 (rank : 15) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DSG2_MOUSE
|
||||||
| NC score | 0.013799 (rank : 16) | θ value | 0.62314 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55111, Q8K069, Q8R517 | Gene names | Dsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor. | |||||
|
ZN700_HUMAN
|
||||||
| NC score | 0.011948 (rank : 17) | θ value | 0.0330416 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M5 | Gene names | ZNF700 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 700. | |||||
|
ZN440_HUMAN
|
||||||
| NC score | 0.006692 (rank : 18) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYI8, Q8N1R9 | Gene names | ZNF440 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 440. | |||||
|
ZN611_HUMAN
|
||||||
| NC score | 0.005193 (rank : 19) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N823, Q69YG9 | Gene names | ZNF611 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 611. | |||||