Please be patient as the page loads
|
PAP2_HUMAN
|
||||||
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PAP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
PAPPA_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.947119 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PAPPA_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.951476 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
DAF2_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 4) | NC score | 0.574623 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |
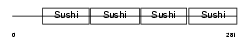
|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
DAF1_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 5) | NC score | 0.567521 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
LYAM3_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 6) | NC score | 0.494863 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P16109, Q8IVD1 | Gene names | SELP, GMRP | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
DAF_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 7) | NC score | 0.555361 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |
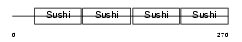
|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
LYAM2_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 8) | NC score | 0.481532 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
LYAM2_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 9) | NC score | 0.471087 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
LYAM3_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 10) | NC score | 0.482755 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q01102 | Gene names | Selp, Grmp | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
CR1_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 11) | NC score | 0.513192 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P17927, Q16745, Q9UQV2 | Gene names | CR1, C3BR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 1 precursor (C3b/C4b receptor) (CD35 antigen). | |||||
|
CSMD1_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 12) | NC score | 0.400338 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CFAH_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 13) | NC score | 0.480086 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P08603, P78435, Q14570, Q38G77, Q5TFM3, Q8N708, Q9NU86 | Gene names | CFH, HF, HF1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (H factor 1). | |||||
|
CFAH_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 14) | NC score | 0.495166 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P06909 | Gene names | Cfh, Hf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (Protein beta-1-H). | |||||
|
CR2_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 15) | NC score | 0.504306 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P19070 | Gene names | Cr2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (CD21 antigen). | |||||
|
CR2_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 16) | NC score | 0.505508 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P20023, Q13866, Q14212 | Gene names | CR2, C3DR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (Epstein-Barr virus receptor) (EBV receptor) (CD21 antigen). | |||||
|
CSMD3_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 17) | NC score | 0.395823 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
ZP3R_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 18) | NC score | 0.504948 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60736 | Gene names | Zp3r, Sp56 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 receptor precursor (Sperm fertilization protein 56) (sp56). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 19) | NC score | 0.386791 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 20) | NC score | 0.396419 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
F13B_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 21) | NC score | 0.472587 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
FHR4_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 22) | NC score | 0.401258 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92496, Q9UJY6 | Gene names | CFHL4, FHR4 | |||
|
Domain Architecture |
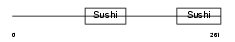
|
|||||
| Description | Complement factor H-related protein 4 precursor (FHR-4). | |||||
|
SRPX_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 23) | NC score | 0.480382 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P78539, Q99652, Q99913 | Gene names | SRPX, ETX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sushi repeat-containing protein SRPX precursor. | |||||
|
FHR3_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 24) | NC score | 0.410973 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q02985, Q9UJ16 | Gene names | CFHL3, FHR3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 3 precursor (FHR-3) (H factor-like protein 3) (DOWN16). | |||||
|
C4BP_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 25) | NC score | 0.495982 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P04003 | Gene names | C4BPA, C4BP | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein alpha chain precursor (C4bp) (Proline-rich protein) (PRP). | |||||
|
F13B_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 26) | NC score | 0.470038 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P05160 | Gene names | F13B | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain) (Fibrin- stabilizing factor B subunit). | |||||
|
SUSD4_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 27) | NC score | 0.449588 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
C4BB_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 28) | NC score | 0.472784 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P20851 | Gene names | C4BPB | |||
|
Domain Architecture |
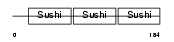
|
|||||
| Description | C4b-binding protein beta chain precursor. | |||||
|
C4BP_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 29) | NC score | 0.483562 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P08607 | Gene names | C4bpa, C4bp | |||
|
Domain Architecture |
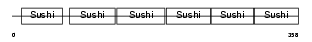
|
|||||
| Description | C4b-binding protein precursor (C4bp). | |||||
|
SUSD1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 30) | NC score | 0.240918 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
SRPX_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 31) | NC score | 0.480026 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9R0M3, Q9R0M2 | Gene names | Srpx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi-repeat-containing protein SRPX precursor (DRS protein). | |||||
|
APOH_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 32) | NC score | 0.490783 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01339 | Gene names | Apoh, B2gp1 | |||
|
Domain Architecture |
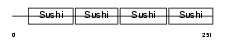
|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor). | |||||
|
SUSD4_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 33) | NC score | 0.425952 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
CO2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 34) | NC score | 0.252878 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
LYAM1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 35) | NC score | 0.370350 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |
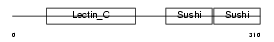
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 36) | NC score | 0.080892 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
FHR2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 37) | NC score | 0.413320 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P36980, Q14310 | Gene names | CFHL2, FHR2, HFL3 | |||
|
Domain Architecture |
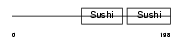
|
|||||
| Description | Complement factor H-related protein 2 precursor (FHR-2) (H factor-like protein 2) (H factor-like 3) (DDESK59). | |||||
|
ADAM7_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 38) | NC score | 0.032987 (rank : 168) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35227 | Gene names | Adam7 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 7 precursor (A disintegrin and metalloproteinase domain 7). | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 39) | NC score | 0.078017 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 40) | NC score | 0.080203 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
TNR14_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 41) | NC score | 0.083923 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |
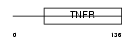
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
FHR1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 42) | NC score | 0.430880 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03591, Q9UJ17 | Gene names | CFHL1, CFHL, FHR1, HFL1 | |||
|
Domain Architecture |
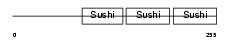
|
|||||
| Description | Complement factor H-related protein 1 precursor (FHR-1) (H factor-like protein 1) (H-factor-like 1) (H36). | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 43) | NC score | 0.087771 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
FHR5_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.432495 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BXR6 | Gene names | CFHL5, FHR5 | |||
|
Domain Architecture |
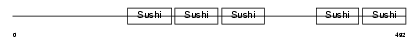
|
|||||
| Description | Complement factor H-related protein 5 precursor (FHR-5). | |||||
|
GNPTA_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 45) | NC score | 0.132298 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q69ZN6, Q3U3K6, Q3US34 | Gene names | Gnptab, Gnpta, Kiaa1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 46) | NC score | 0.083642 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 47) | NC score | 0.084779 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 48) | NC score | 0.085692 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 49) | NC score | 0.075479 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 50) | NC score | 0.055235 (rank : 142) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
TNR8_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 51) | NC score | 0.058785 (rank : 122) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |
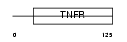
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
LYAM1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 52) | NC score | 0.355973 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |
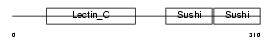
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 53) | NC score | 0.081247 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
CRRY_MOUSE
|
||||||
| θ value | 0.125558 (rank : 54) | NC score | 0.484449 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q64735, Q3TV30, Q58E68, Q61447, Q61449, Q8CF59, Q8K328 | Gene names | Crry, Cry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement regulatory protein Crry precursor (Protein p65). | |||||
|
PCSK5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 55) | NC score | 0.050669 (rank : 161) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
APOH_HUMAN
|
||||||
| θ value | 0.163984 (rank : 56) | NC score | 0.442473 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P02749, Q9UCN7 | Gene names | APOH, B2G1 | |||
|
Domain Architecture |
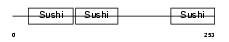
|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor) (Anticardiolipin cofactor). | |||||
|
C1R_HUMAN
|
||||||
| θ value | 0.163984 (rank : 57) | NC score | 0.078958 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
GNPTA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 58) | NC score | 0.123810 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3T906, Q3ZQK2, Q6IPW5, Q86TQ2, Q96N13, Q9ULL2 | Gene names | GNPTAB, GNPTA, KIAA1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
ADM1A_MOUSE
|
||||||
| θ value | 0.279714 (rank : 59) | NC score | 0.030983 (rank : 172) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60813, Q60617, Q80WR7, Q8R533 | Gene names | Adam1a, Adam1, Ftna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 1a precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 1a) (Fertilin alpha 1a subunit). | |||||
|
ADA17_HUMAN
|
||||||
| θ value | 0.365318 (rank : 60) | NC score | 0.041074 (rank : 163) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||
|
ATS7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 61) | NC score | 0.019955 (rank : 176) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKP4 | Gene names | ADAMTS7 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-7 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 7) (ADAM-TS 7) (ADAM-TS7). | |||||
|
MASP2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 62) | NC score | 0.122173 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
LAMC3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.051823 (rank : 158) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
ADAM8_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.033103 (rank : 167) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78325 | Gene names | ADAM8, MS2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (CD156a antigen) (CD156). | |||||
|
ADA10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.037239 (rank : 166) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35598 | Gene names | Adam10, Kuz, Madm | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 10 precursor (EC 3.4.24.81) (A disintegrin and metalloproteinase domain 10) (Mammalian disintegrin-metalloprotease) (Kuzbanian protein homolog). | |||||
|
JAG1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.072386 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.072053 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.087502 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
TNAP3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.028335 (rank : 173) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60769 | Gene names | Tnfaip3, Tnfip3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
CO2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.192992 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
CS1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.094178 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
CS1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.095836 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.045538 (rank : 162) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.079946 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
USH2A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.039285 (rank : 165) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
FAAA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.026272 (rank : 175) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35505 | Gene names | Fah | |||
|
Domain Architecture |

|
|||||
| Description | Fumarylacetoacetase (EC 3.7.1.2) (Fumarylacetoacetate hydrolase) (Beta-diketonase) (FAA). | |||||
|
HXA5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.006494 (rank : 182) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09021, Q91VQ5 | Gene names | Hoxa5, Hox-1.3, Hoxa-5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1.3) (M2). | |||||
|
COLQ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.032592 (rank : 170) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y215, Q6DK18, Q6YH18, Q6YH19, Q6YH20, Q6YH21, Q9NP18, Q9NP19, Q9NP20, Q9NP21, Q9NP22, Q9NP23, Q9NP24, Q9UP88 | Gene names | COLQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acetylcholinesterase collagenic tail peptide precursor (AChE Q subunit) (Acetylcholinesterase-associated collagen). | |||||
|
ADA25_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.027184 (rank : 174) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R159, Q9D4E4 | Gene names | Adam25 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 25 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 25) (Testase 2). | |||||
|
NELL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.051543 (rank : 160) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.068369 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
ZN548_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | -0.002822 (rank : 188) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEK5, Q96M05 | Gene names | ZNF548 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 548. | |||||
|
APR3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.040036 (rank : 164) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PGD0, Q3UAH4, Q810Q3, Q9DD14 | Gene names | Apr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-related protein 3 precursor (APR-3). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.011406 (rank : 180) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
HXA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.005175 (rank : 183) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20719, O43367, Q96CY6 | Gene names | HOXA5, HOX1C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1C). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.008754 (rank : 181) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
T2EA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.019080 (rank : 178) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29083, Q16103 | Gene names | GTF2E1, TF2E1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE 56 kDa subunit). | |||||
|
CFAB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.211648 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.072967 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.324446 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
STAB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.055622 (rank : 140) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | -0.002290 (rank : 186) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
C1S_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.111049 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
CO8A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.015467 (rank : 179) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P07357, Q13668, Q9H130 | Gene names | C8A | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.004958 (rank : 184) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
FBN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.065547 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.051916 (rank : 157) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
GALT6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.003157 (rank : 185) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
ITB6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.030997 (rank : 171) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
JAG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.071647 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
ADA10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.032665 (rank : 169) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14672, Q10742, Q92650 | Gene names | ADAM10, KUZ, MADM | |||
|
Domain Architecture |
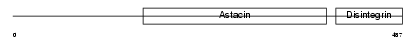
|
|||||
| Description | ADAM 10 precursor (EC 3.4.24.81) (A disintegrin and metalloproteinase domain 10) (Mammalian disintegrin-metalloprotease) (Kuzbanian protein homolog) (CDw156c antigen). | |||||
|
CO6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.166071 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.052949 (rank : 152) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
MCP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.413439 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88174, Q9R0R9 | Gene names | Cd46, Mcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane cofactor protein precursor (CD46 antigen). | |||||
|
TNAP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.019710 (rank : 177) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
ZN444_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | -0.002752 (rank : 187) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N0Y2, Q8TEQ9, Q8WU35, Q9NUU1 | Gene names | ZNF444, EZF2, ZSCAN17 | |||
|
Domain Architecture |
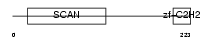
|
|||||
| Description | Zinc finger protein 444 (Endothelial zinc finger protein 2) (EZF-2) (Zinc finger and SCAN domain-containing protein 17). | |||||
|
ATRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.060402 (rank : 117) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.058327 (rank : 124) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
BMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.057467 (rank : 130) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.057709 (rank : 126) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
C1QR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.056270 (rank : 136) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O89103, Q3U2X0 | Gene names | Cd93, Aa4, C1qr1, C1qrp, Ly68 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (Cell surface antigen AA4) (Lymphocyte antigen 68). | |||||
|
C1RA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.074678 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.074693 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
CFAB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.207030 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P00751, O15006, Q29944, Q96HX6, Q9BTF5, Q9BX92 | Gene names | CFB, BF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) (Properdin factor B) (Glycine-rich beta glycoprotein) (GBG) (PBF2) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CO7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.174109 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
CRUM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.057237 (rank : 134) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.055015 (rank : 143) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.055843 (rank : 138) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.082258 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.082239 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.093354 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.098295 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.085595 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.079817 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
DLK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.053305 (rank : 149) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052973 (rank : 151) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.058905 (rank : 121) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.058961 (rank : 120) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.052454 (rank : 154) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052779 (rank : 153) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.051665 (rank : 159) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |
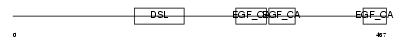
|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.054444 (rank : 144) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DNER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.058312 (rank : 125) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
DNER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.057459 (rank : 131) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
EGFL9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.053367 (rank : 148) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
EGFL9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.053014 (rank : 150) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
FBLN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.052234 (rank : 155) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P23142, P23143, P23144, P37888, Q5TIC4, Q8TBH8, Q9HBQ5, Q9UC21, Q9UGR4, Q9UH41 | Gene names | FBLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor. | |||||
|
FBLN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.057580 (rank : 127) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.055476 (rank : 141) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.057350 (rank : 132) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.065292 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.061554 (rank : 114) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.064298 (rank : 109) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.060443 (rank : 116) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
GABR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.130523 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.129719 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
IL2RA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.064146 (rank : 110) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P01589 | Gene names | IL2RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor alpha chain precursor (IL-2 receptor alpha subunit) (IL-2-RA) (IL2-RA) (p55) (TAC antigen) (CD25 antigen). | |||||
|
JAG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.060647 (rank : 115) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.061699 (rank : 113) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
K0247_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.134363 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92537 | Gene names | KIAA0247 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0247 precursor. | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.072800 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR102_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.077774 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR104_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.087772 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.074028 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR107_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.088714 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR108_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.064121 (rank : 111) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR109_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.071850 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.072129 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.056102 (rank : 137) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
LAMA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.052232 (rank : 156) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.054079 (rank : 146) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.053904 (rank : 147) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.066739 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LTBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.057512 (rank : 129) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.059488 (rank : 119) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.068738 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MASP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.092255 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.085035 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.101836 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MCP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.402256 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15529, Q15429, Q53GV9, Q5HY94, Q5VWS6, Q5VWS7, Q5VWS8, Q5VWS9, Q5VWT0, Q5VWT1, Q5VWT2, Q6N0A1, Q7Z3R5, Q9NNW2, Q9NNW3, Q9NNW4 | Gene names | CD46, MCP | |||
|
Domain Architecture |

|
|||||
| Description | Membrane cofactor protein precursor (Trophoblast leukocyte common antigen) (TLX) (CD46 antigen). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.086334 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
PERT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.085880 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P07202, P09934, P09935, Q8IUL0, Q8NF94, Q8NF95, Q8NF96, Q8NF97, Q8TCI9 | Gene names | TPO | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
PERT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.095373 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
SCUB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.055653 (rank : 139) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SCUB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.054356 (rank : 145) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SREC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.065954 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.082829 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
SREC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.063494 (rank : 112) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
STAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.056654 (rank : 135) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
SUSD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.170504 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
SUSD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.182206 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
SUSD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.087451 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96L08, Q49AA6, Q6UXV7 | Gene names | SUSD3, UNQ9387/PRO34275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
SUSD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.155971 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D176, Q80XL5, Q9CYV1, Q9DA86 | Gene names | Susd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
TLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.057298 (rank : 133) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.059691 (rank : 118) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.057522 (rank : 128) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.058641 (rank : 123) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.080228 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
PAP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
PAPPA_MOUSE
|
||||||
| NC score | 0.951476 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PAPPA_HUMAN
|
||||||
| NC score | 0.947119 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
DAF2_MOUSE
|
||||||
| NC score | 0.574623 (rank : 4) | θ value | 5.8054e-15 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
DAF1_MOUSE
|
||||||
| NC score | 0.567521 (rank : 5) | θ value | 2.20605e-14 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
DAF_HUMAN
|
||||||
| NC score | 0.555361 (rank : 6) | θ value | 1.58096e-12 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
CR1_HUMAN
|
||||||
| NC score | 0.513192 (rank : 7) | θ value | 5.62301e-10 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P17927, Q16745, Q9UQV2 | Gene names | CR1, C3BR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 1 precursor (C3b/C4b receptor) (CD35 antigen). | |||||
|
CR2_HUMAN
|
||||||
| NC score | 0.505508 (rank : 8) | θ value | 1.80886e-08 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P20023, Q13866, Q14212 | Gene names | CR2, C3DR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (Epstein-Barr virus receptor) (EBV receptor) (CD21 antigen). | |||||
|
ZP3R_MOUSE
|
||||||
| NC score | 0.504948 (rank : 9) | θ value | 7.59969e-07 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60736 | Gene names | Zp3r, Sp56 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 receptor precursor (Sperm fertilization protein 56) (sp56). | |||||
|
CR2_MOUSE
|
||||||
| NC score | 0.504306 (rank : 10) | θ value | 2.79066e-09 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P19070 | Gene names | Cr2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (CD21 antigen). | |||||
|
C4BP_HUMAN
|
||||||
| NC score | 0.495982 (rank : 11) | θ value | 4.1701e-05 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P04003 | Gene names | C4BPA, C4BP | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein alpha chain precursor (C4bp) (Proline-rich protein) (PRP). | |||||
|
CFAH_MOUSE
|
||||||
| NC score | 0.495166 (rank : 12) | θ value | 2.13673e-09 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P06909 | Gene names | Cfh, Hf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (Protein beta-1-H). | |||||
|
LYAM3_HUMAN
|
||||||
| NC score | 0.494863 (rank : 13) | θ value | 1.86753e-13 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P16109, Q8IVD1 | Gene names | SELP, GMRP | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
APOH_MOUSE
|
||||||
| NC score | 0.490783 (rank : 14) | θ value | 0.00020696 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01339 | Gene names | Apoh, B2gp1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor). | |||||
|
CRRY_MOUSE
|
||||||
| NC score | 0.484449 (rank : 15) | θ value | 0.125558 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q64735, Q3TV30, Q58E68, Q61447, Q61449, Q8CF59, Q8K328 | Gene names | Crry, Cry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement regulatory protein Crry precursor (Protein p65). | |||||
|
C4BP_MOUSE
|
||||||
| NC score | 0.483562 (rank : 16) | θ value | 5.44631e-05 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P08607 | Gene names | C4bpa, C4bp | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein precursor (C4bp). | |||||
|
LYAM3_MOUSE
|
||||||
| NC score | 0.482755 (rank : 17) | θ value | 3.89403e-11 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q01102 | Gene names | Selp, Grmp | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
LYAM2_HUMAN
|
||||||
| NC score | 0.481532 (rank : 18) | θ value | 3.52202e-12 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
SRPX_HUMAN
|
||||||
| NC score | 0.480382 (rank : 19) | θ value | 1.09739e-05 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P78539, Q99652, Q99913 | Gene names | SRPX, ETX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sushi repeat-containing protein SRPX precursor. | |||||
|
CFAH_HUMAN
|
||||||
| NC score | 0.480086 (rank : 20) | θ value | 2.13673e-09 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P08603, P78435, Q14570, Q38G77, Q5TFM3, Q8N708, Q9NU86 | Gene names | CFH, HF, HF1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (H factor 1). | |||||
|
SRPX_MOUSE
|
||||||
| NC score | 0.480026 (rank : 21) | θ value | 9.29e-05 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9R0M3, Q9R0M2 | Gene names | Srpx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi-repeat-containing protein SRPX precursor (DRS protein). | |||||
|
C4BB_HUMAN
|
||||||
| NC score | 0.472784 (rank : 22) | θ value | 5.44631e-05 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P20851 | Gene names | C4BPB | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein beta chain precursor. | |||||
|
F13B_MOUSE
|
||||||
| NC score | 0.472587 (rank : 23) | θ value | 2.21117e-06 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
LYAM2_MOUSE
|
||||||
| NC score | 0.471087 (rank : 24) | θ value | 1.74796e-11 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
F13B_HUMAN
|
||||||
| NC score | 0.470038 (rank : 25) | θ value | 4.1701e-05 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P05160 | Gene names | F13B | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain) (Fibrin- stabilizing factor B subunit). | |||||
|
SUSD4_HUMAN
|
||||||
| NC score | 0.449588 (rank : 26) | θ value | 4.1701e-05 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
APOH_HUMAN
|
||||||
| NC score | 0.442473 (rank : 27) | θ value | 0.163984 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P02749, Q9UCN7 | Gene names | APOH, B2G1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor) (Anticardiolipin cofactor). | |||||
|
FHR5_HUMAN
|
||||||
| NC score | 0.432495 (rank : 28) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9BXR6 | Gene names | CFHL5, FHR5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 5 precursor (FHR-5). | |||||
|
FHR1_HUMAN
|
||||||
| NC score | 0.430880 (rank : 29) | θ value | 0.0563607 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03591, Q9UJ17 | Gene names | CFHL1, CFHL, FHR1, HFL1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 1 precursor (FHR-1) (H factor-like protein 1) (H-factor-like 1) (H36). | |||||
|
SUSD4_MOUSE
|
||||||
| NC score | 0.425952 (rank : 30) | θ value | 0.00035302 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
MCP_MOUSE
|
||||||
| NC score | 0.413439 (rank : 31) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88174, Q9R0R9 | Gene names | Cd46, Mcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane cofactor protein precursor (CD46 antigen). | |||||
|
FHR2_HUMAN
|
||||||
| NC score | 0.413320 (rank : 32) | θ value | 0.00869519 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P36980, Q14310 | Gene names | CFHL2, FHR2, HFL3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 2 precursor (FHR-2) (H factor-like protein 2) (H factor-like 3) (DDESK59). | |||||
|
FHR3_HUMAN
|
||||||
| NC score | 0.410973 (rank : 33) | θ value | 3.19293e-05 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q02985, Q9UJ16 | Gene names | CFHL3, FHR3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 3 precursor (FHR-3) (H factor-like protein 3) (DOWN16). | |||||
|
MCP_HUMAN
|
||||||
| NC score | 0.402256 (rank : 34) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15529, Q15429, Q53GV9, Q5HY94, Q5VWS6, Q5VWS7, Q5VWS8, Q5VWS9, Q5VWT0, Q5VWT1, Q5VWT2, Q6N0A1, Q7Z3R5, Q9NNW2, Q9NNW3, Q9NNW4 | Gene names | CD46, MCP | |||
|
Domain Architecture |

|
|||||
| Description | Membrane cofactor protein precursor (Trophoblast leukocyte common antigen) (TLX) (CD46 antigen). | |||||
|
FHR4_HUMAN
|
||||||
| NC score | 0.401258 (rank : 35) | θ value | 4.92598e-06 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q92496, Q9UJY6 | Gene names | CFHL4, FHR4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 4 precursor (FHR-4). | |||||
|
CSMD1_MOUSE
|
||||||
| NC score | 0.400338 (rank : 36) | θ value | 1.63604e-09 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.396419 (rank : 37) | θ value | 2.21117e-06 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CSMD3_HUMAN
|
||||||
| NC score | 0.395823 (rank : 38) | θ value | 3.41135e-07 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.386791 (rank : 39) | θ value | 9.92553e-07 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
LYAM1_MOUSE
|
||||||
| NC score | 0.370350 (rank : 40) | θ value | 0.00228821 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
LYAM1_HUMAN
|
||||||
| NC score | 0.355973 (rank : 41) | θ value | 0.0961366 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.324446 (rank : 42) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
CO2_HUMAN
|
||||||
| NC score | 0.252878 (rank : 43) | θ value | 0.000786445 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
SUSD1_HUMAN
|
||||||
| NC score | 0.240918 (rank : 44) | θ value | 5.44631e-05 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
CFAB_MOUSE
|
||||||
| NC score | 0.211648 (rank : 45) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CFAB_HUMAN
|
||||||
| NC score | 0.207030 (rank : 46) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P00751, O15006, Q29944, Q96HX6, Q9BTF5, Q9BX92 | Gene names | CFB, BF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) (Properdin factor B) (Glycine-rich beta glycoprotein) (GBG) (PBF2) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CO2_MOUSE
|
||||||
| NC score | 0.192992 (rank : 47) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
SUSD2_MOUSE
|
||||||
| NC score | 0.182206 (rank : 48) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
CO7_HUMAN
|
||||||
| NC score | 0.174109 (rank : 49) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
SUSD2_HUMAN
|
||||||
| NC score | 0.170504 (rank : 50) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
CO6_HUMAN
|
||||||
| NC score | 0.166071 (rank : 51) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
SUSD3_MOUSE
|
||||||
| NC score | 0.155971 (rank : 52) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D176, Q80XL5, Q9CYV1, Q9DA86 | Gene names | Susd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
K0247_HUMAN
|
||||||
| NC score | 0.134363 (rank : 53) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92537 | Gene names | KIAA0247 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0247 precursor. | |||||
|
GNPTA_MOUSE
|
||||||
| NC score | 0.132298 (rank : 54) | θ value | 0.0736092 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q69ZN6, Q3U3K6, Q3US34 | Gene names | Gnptab, Gnpta, Kiaa1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.130523 (rank : 55) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.129719 (rank : 56) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GNPTA_HUMAN
|
||||||
| NC score | 0.123810 (rank : 57) | θ value | 0.21417 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3T906, Q3ZQK2, Q6IPW5, Q86TQ2, Q96N13, Q9ULL2 | Gene names | GNPTAB, GNPTA, KIAA1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
MASP2_MOUSE
|
||||||
| NC score | 0.122173 (rank : 58) | θ value | 0.365318 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
C1S_HUMAN
|
||||||
| NC score | 0.111049 (rank : 59) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.101836 (rank : 60) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.098295 (rank : 61) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CS1B_MOUSE
|
||||||
| NC score | 0.095836 (rank : 62) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
PERT_MOUSE
|
||||||
| NC score | 0.095373 (rank : 63) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
CS1A_MOUSE
|
||||||
| NC score | 0.094178 (rank : 64) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.093354 (rank : 65) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.092255 (rank : 66) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.088714 (rank : 67) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.087772 (rank : 68) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.087771 (rank : 69) | θ value | 0.0563607 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.087502 (rank : 70) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
SUSD3_HUMAN
|
||||||
| NC score | 0.087451 (rank : 71) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96L08, Q49AA6, Q6UXV7 | Gene names | SUSD3, UNQ9387/PRO34275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.086334 (rank : 72) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
PERT_HUMAN
|
||||||
| NC score | 0.085880 (rank : 73) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P07202, P09934, P09935, Q8IUL0, Q8NF94, Q8NF95, Q8NF96, Q8NF97, Q8TCI9 | Gene names | TPO | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.085692 (rank : 74) | θ value | 0.0736092 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.085595 (rank : 75) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
MASP1_MOUSE
|
||||||
| NC score | 0.085035 (rank : 76) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.084779 (rank : 77) | θ value | 0.0736092 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
TNR14_HUMAN
|
||||||
| NC score | 0.083923 (rank : 78) | θ value | 0.0431538 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.083642 (rank : 79) | θ value | 0.0736092 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.082829 (rank : 80) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.082258 (rank : 81) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.082239 (rank : 82) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.081247 (rank : 83) | θ value | 0.0961366 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.080892 (rank : 84) | θ value | 0.00228821 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.080228 (rank : 85) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.080203 (rank : 86) | θ value | 0.0431538 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.079946 (rank : 87) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.079817 (rank : 88) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
C1R_HUMAN
|
||||||
| NC score | 0.078958 (rank : 89) | θ value | 0.163984 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.078017 (rank : 90) | θ value | 0.0330416 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.077774 (rank : 91) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.075479 (rank : 92) | θ value | 0.0736092 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
C1RB_MOUSE
|
||||||
| NC score | 0.074693 (rank : 93) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
C1RA_MOUSE
|
||||||
| NC score | 0.074678 (rank : 94) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.074028 (rank : 95) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.072967 (rank : 96) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.072800 (rank : 97) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.072386 (rank : 98) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.072129 (rank : 99) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.072053 (rank : 100) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.071850 (rank : 101) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.071647 (rank : 102) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.068738 (rank : 103) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.068369 (rank : 104) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.066739 (rank : 105) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.065954 (rank : 106) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
FBN1_MOUSE
|
||||||
| NC score | 0.065547 (rank : 107) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.065292 (rank : 108) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.064298 (rank : 109) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
IL2RA_HUMAN
|
||||||
| NC score | 0.064146 (rank : 110) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P01589 | Gene names | IL2RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor alpha chain precursor (IL-2 receptor alpha subunit) (IL-2-RA) (IL2-RA) (p55) (TAC antigen) (CD25 antigen). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.064121 (rank : 111) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.063494 (rank : 112) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.061699 (rank : 113) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.061554 (rank : 114) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.060647 (rank : 115) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.060443 (rank : 116) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.060402 (rank : 117) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
TLL1_MOUSE
|
||||||
| NC score | 0.059691 (rank : 118) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
LTBP3_MOUSE
|
||||||
| NC score | 0.059488 (rank : 119) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.058961 (rank : 120) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.058905 (rank : 121) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
TNR8_MOUSE
|
||||||
| NC score | 0.058785 (rank : 122) | θ value | 0.0736092 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
TLL2_MOUSE
|
||||||
| NC score | 0.058641 (rank : 123) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.058327 (rank : 124) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
DNER_HUMAN
|
||||||
| NC score | 0.058312 (rank : 125) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
BMP1_MOUSE
|
||||||
| NC score | 0.057709 (rank : 126) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
FBLN1_MOUSE
|
||||||
| NC score | 0.057580 (rank : 127) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
TLL2_HUMAN
|
||||||
| NC score | 0.057522 (rank : 128) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
LTBP3_HUMAN
|
||||||
| NC score | 0.057512 (rank : 129) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
BMP1_HUMAN
|
||||||
| NC score | 0.057467 (rank : 130) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
DNER_MOUSE
|
||||||
| NC score | 0.057459 (rank : 131) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8JZM4, Q3U697, Q8R226, Q8R4T6, Q8VD97 | Gene names | Dner, Bet, Bret | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor (Brain EGF repeat-containing transmembrane protein). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.057350 (rank : 132) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
TLL1_HUMAN
|
||||||
| NC score | 0.057298 (rank : 133) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
CRUM1_HUMAN
|
||||||
| NC score | 0.057237 (rank : 134) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.056654 (rank : 135) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
C1QR1_MOUSE
|
||||||
| NC score | 0.056270 (rank : 136) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O89103, Q3U2X0 | Gene names | Cd93, Aa4, C1qr1, C1qrp, Ly68 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (Cell surface antigen AA4) (Lymphocyte antigen 68). | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.056102 (rank : 137) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
CRUM2_HUMAN
|
||||||
| NC score | 0.055843 (rank : 138) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
SCUB3_HUMAN
|
||||||
| NC score | 0.055653 (rank : 139) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.055622 (rank : 140) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.055476 (rank : 141) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.055235 (rank : 142) | θ value | 0.0736092 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| NC score | 0.055015 (rank : 143) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.054444 (rank : 144) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
SCUB3_MOUSE
|
||||||
| NC score | 0.054356 (rank : 145) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.054079 (rank : 146) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| NC score | 0.053904 (rank : 147) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
EGFL9_HUMAN
|
||||||
| NC score | 0.053367 (rank : 148) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
DLK_HUMAN
|
||||||
| NC score | 0.053305 (rank : 149) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
EGFL9_MOUSE
|
||||||
| NC score | 0.053014 (rank : 150) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1E3, Q9QYP3 | Gene names | Egfl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9) (Endothelial cell-specific protein S-1). | |||||
|
DLK_MOUSE
|
||||||
| NC score | 0.052973 (rank : 151) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.052949 (rank : 152) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.052779 (rank : 153) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.052454 (rank : 154) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
FBLN1_HUMAN
|
||||||
| NC score | 0.052234 (rank : 155) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P23142, P23143, P23144, P37888, Q5TIC4, Q8TBH8, Q9HBQ5, Q9UC21, Q9UGR4, Q9UH41 | Gene names | FBLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor. | |||||
|
LAMA5_HUMAN
|
||||||
| NC score | 0.052232 (rank : 156) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.051916 (rank : 157) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
LAMC3_HUMAN
|
||||||
| NC score | 0.051823 (rank : 158) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
DLL4_HUMAN
|
||||||
| NC score | 0.051665 (rank : 159) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
NELL2_MOUSE
|
||||||
| NC score | 0.051543 (rank : 160) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
PCSK5_HUMAN
|
||||||
| NC score | 0.050669 (rank : 161) | θ value | 0.125558 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.045538 (rank : 162) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
ADA17_HUMAN
|
||||||
| NC score | 0.041074 (rank : 163) | θ value | 0.365318 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||
|
APR3_MOUSE
|
||||||
| NC score | 0.040036 (rank : 164) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PGD0, Q3UAH4, Q810Q3, Q9DD14 | Gene names | Apr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-related protein 3 precursor (APR-3). | |||||
|
USH2A_HUMAN
|
||||||
| NC score | 0.039285 (rank : 165) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
ADA10_MOUSE
|
||||||
| NC score | 0.037239 (rank : 166) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35598 | Gene names | Adam10, Kuz, Madm | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 10 precursor (EC 3.4.24.81) (A disintegrin and metalloproteinase domain 10) (Mammalian disintegrin-metalloprotease) (Kuzbanian protein homolog). | |||||
|
ADAM8_HUMAN
|
||||||
| NC score | 0.033103 (rank : 167) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78325 | Gene names | ADAM8, MS2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (CD156a antigen) (CD156). | |||||
|
ADAM7_MOUSE
|
||||||
| NC score | 0.032987 (rank : 168) | θ value | 0.0330416 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35227 | Gene names | Adam7 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 7 precursor (A disintegrin and metalloproteinase domain 7). | |||||
|
ADA10_HUMAN
|
||||||
| NC score | 0.032665 (rank : 169) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14672, Q10742, Q92650 | Gene names | ADAM10, KUZ, MADM | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 10 precursor (EC 3.4.24.81) (A disintegrin and metalloproteinase domain 10) (Mammalian disintegrin-metalloprotease) (Kuzbanian protein homolog) (CDw156c antigen). | |||||
|
COLQ_HUMAN
|
||||||
| NC score | 0.032592 (rank : 170) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y215, Q6DK18, Q6YH18, Q6YH19, Q6YH20, Q6YH21, Q9NP18, Q9NP19, Q9NP20, Q9NP21, Q9NP22, Q9NP23, Q9NP24, Q9UP88 | Gene names | COLQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acetylcholinesterase collagenic tail peptide precursor (AChE Q subunit) (Acetylcholinesterase-associated collagen). | |||||
|
ITB6_MOUSE
|
||||||
| NC score | 0.030997 (rank : 171) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ADM1A_MOUSE
|
||||||
| NC score | 0.030983 (rank : 172) | θ value | 0.279714 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60813, Q60617, Q80WR7, Q8R533 | Gene names | Adam1a, Adam1, Ftna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 1a precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 1a) (Fertilin alpha 1a subunit). | |||||
|
TNAP3_MOUSE
|
||||||
| NC score | 0.028335 (rank : 173) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60769 | Gene names | Tnfaip3, Tnfip3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
ADA25_MOUSE
|
||||||
| NC score | 0.027184 (rank : 174) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R159, Q9D4E4 | Gene names | Adam25 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 25 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 25) (Testase 2). | |||||
|
FAAA_MOUSE
|
||||||
| NC score | 0.026272 (rank : 175) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35505 | Gene names | Fah | |||
|
Domain Architecture |

|
|||||
| Description | Fumarylacetoacetase (EC 3.7.1.2) (Fumarylacetoacetate hydrolase) (Beta-diketonase) (FAA). | |||||
|
ATS7_HUMAN
|
||||||
| NC score | 0.019955 (rank : 176) | θ value | 0.365318 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKP4 | Gene names | ADAMTS7 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-7 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 7) (ADAM-TS 7) (ADAM-TS7). | |||||
|
TNAP3_HUMAN
|
||||||
| NC score | 0.019710 (rank : 177) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
T2EA_HUMAN
|
||||||
| NC score | 0.019080 (rank : 178) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29083, Q16103 | Gene names | GTF2E1, TF2E1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIE subunit alpha (TFIIE-alpha) (General transcription factor IIE 56 kDa subunit). | |||||
|
CO8A_HUMAN
|
||||||
| NC score | 0.015467 (rank : 179) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P07357, Q13668, Q9H130 | Gene names | C8A | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.011406 (rank : 180) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.008754 (rank : 181) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
HXA5_MOUSE
|
||||||
| NC score | 0.006494 (rank : 182) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09021, Q91VQ5 | Gene names | Hoxa5, Hox-1.3, Hoxa-5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1.3) (M2). | |||||
|
HXA5_HUMAN
|
||||||
| NC score | 0.005175 (rank : 183) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20719, O43367, Q96CY6 | Gene names | HOXA5, HOX1C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1C). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.004958 (rank : 184) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
GALT6_MOUSE
|
||||||
| NC score | 0.003157 (rank : 185) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C7U7, Q3TWF0, Q8CED2, Q9QZ16 | Gene names | Galnt6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 6 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 6) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 6) (Polypeptide GalNAc transferase 6) (GalNAc-T6) (pp-GaNTase 6). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.002290 (rank : 186) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZN444_HUMAN
|
||||||
| NC score | -0.002752 (rank : 187) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N0Y2, Q8TEQ9, Q8WU35, Q9NUU1 | Gene names | ZNF444, EZF2, ZSCAN17 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 444 (Endothelial zinc finger protein 2) (EZF-2) (Zinc finger and SCAN domain-containing protein 17). | |||||
|
ZN548_HUMAN
|
||||||
| NC score | -0.002822 (rank : 188) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEK5, Q96M05 | Gene names | ZNF548 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 548. | |||||