Please be patient as the page loads
|
DAF_HUMAN
|
||||||
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |
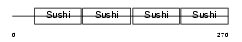
|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DAF_HUMAN
|
||||||
| θ value | 3.0341e-181 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
DAF1_MOUSE
|
||||||
| θ value | 2.37764e-101 (rank : 2) | NC score | 0.965322 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
DAF2_MOUSE
|
||||||
| θ value | 2.81514e-94 (rank : 3) | NC score | 0.974351 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |
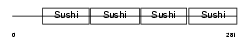
|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
CR1_HUMAN
|
||||||
| θ value | 1.2411e-41 (rank : 4) | NC score | 0.910123 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P17927, Q16745, Q9UQV2 | Gene names | CR1, C3BR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 1 precursor (C3b/C4b receptor) (CD35 antigen). | |||||
|
CRRY_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 5) | NC score | 0.939843 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q64735, Q3TV30, Q58E68, Q61447, Q61449, Q8CF59, Q8K328 | Gene names | Crry, Cry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement regulatory protein Crry precursor (Protein p65). | |||||
|
C4BP_HUMAN
|
||||||
| θ value | 1.85459e-37 (rank : 6) | NC score | 0.924231 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P04003 | Gene names | C4BPA, C4BP | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein alpha chain precursor (C4bp) (Proline-rich protein) (PRP). | |||||
|
ZP3R_MOUSE
|
||||||
| θ value | 8.61488e-35 (rank : 7) | NC score | 0.927491 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q60736 | Gene names | Zp3r, Sp56 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 receptor precursor (Sperm fertilization protein 56) (sp56). | |||||
|
C4BP_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 8) | NC score | 0.924905 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P08607 | Gene names | C4bpa, C4bp | |||
|
Domain Architecture |
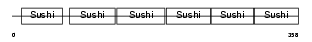
|
|||||
| Description | C4b-binding protein precursor (C4bp). | |||||
|
CR2_MOUSE
|
||||||
| θ value | 1.4233e-29 (rank : 9) | NC score | 0.892569 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P19070 | Gene names | Cr2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (CD21 antigen). | |||||
|
CR2_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 10) | NC score | 0.889103 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P20023, Q13866, Q14212 | Gene names | CR2, C3DR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (Epstein-Barr virus receptor) (EBV receptor) (CD21 antigen). | |||||
|
CSMD3_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 11) | NC score | 0.695266 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 12) | NC score | 0.689352 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
MCP_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 13) | NC score | 0.899960 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P15529, Q15429, Q53GV9, Q5HY94, Q5VWS6, Q5VWS7, Q5VWS8, Q5VWS9, Q5VWT0, Q5VWT1, Q5VWT2, Q6N0A1, Q7Z3R5, Q9NNW2, Q9NNW3, Q9NNW4 | Gene names | CD46, MCP | |||
|
Domain Architecture |

|
|||||
| Description | Membrane cofactor protein precursor (Trophoblast leukocyte common antigen) (TLX) (CD46 antigen). | |||||
|
CSMD1_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 14) | NC score | 0.683805 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CFAH_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 15) | NC score | 0.815744 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P06909 | Gene names | Cfh, Hf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (Protein beta-1-H). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 16) | NC score | 0.673297 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
LYAM3_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 17) | NC score | 0.711742 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q01102 | Gene names | Selp, Grmp | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
MCP_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 18) | NC score | 0.885526 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88174, Q9R0R9 | Gene names | Cd46, Mcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane cofactor protein precursor (CD46 antigen). | |||||
|
LYAM2_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 19) | NC score | 0.697776 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
APOH_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 20) | NC score | 0.854769 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P02749, Q9UCN7 | Gene names | APOH, B2G1 | |||
|
Domain Architecture |
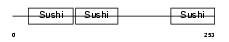
|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor) (Anticardiolipin cofactor). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 21) | NC score | 0.678097 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
LYAM3_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 22) | NC score | 0.715699 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P16109, Q8IVD1 | Gene names | SELP, GMRP | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
CFAH_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 23) | NC score | 0.803044 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P08603, P78435, Q14570, Q38G77, Q5TFM3, Q8N708, Q9NU86 | Gene names | CFH, HF, HF1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (H factor 1). | |||||
|
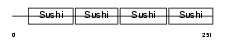
APOH_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 24) | NC score | 0.861599 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q01339 | Gene names | Apoh, B2gp1 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor). | |||||
|
LYAM2_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 25) | NC score | 0.685390 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
F13B_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 26) | NC score | 0.806945 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
PAP2_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 27) | NC score | 0.555361 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
F13B_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 28) | NC score | 0.796676 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P05160 | Gene names | F13B | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain) (Fibrin- stabilizing factor B subunit). | |||||
|
FHR1_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 29) | NC score | 0.715218 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q03591, Q9UJ17 | Gene names | CFHL1, CFHL, FHR1, HFL1 | |||
|
Domain Architecture |
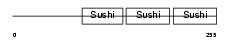
|
|||||
| Description | Complement factor H-related protein 1 precursor (FHR-1) (H factor-like protein 1) (H-factor-like 1) (H36). | |||||
|
PAPPA_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 30) | NC score | 0.603424 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
C4BB_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 31) | NC score | 0.846104 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P20851 | Gene names | C4BPB | |||
|
Domain Architecture |
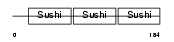
|
|||||
| Description | C4b-binding protein beta chain precursor. | |||||
|
CFAB_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 32) | NC score | 0.463268 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P00751, O15006, Q29944, Q96HX6, Q9BTF5, Q9BX92 | Gene names | CFB, BF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) (Properdin factor B) (Glycine-rich beta glycoprotein) (GBG) (PBF2) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CFAB_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 33) | NC score | 0.466238 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
FHR2_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 34) | NC score | 0.661551 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P36980, Q14310 | Gene names | CFHL2, FHR2, HFL3 | |||
|
Domain Architecture |
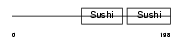
|
|||||
| Description | Complement factor H-related protein 2 precursor (FHR-2) (H factor-like protein 2) (H factor-like 3) (DDESK59). | |||||
|
PAPPA_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 35) | NC score | 0.607923 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
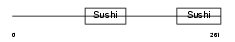
FHR4_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 36) | NC score | 0.658303 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92496, Q9UJY6 | Gene names | CFHL4, FHR4 | |||
|
Domain Architecture |
|
|||||
| Description | Complement factor H-related protein 4 precursor (FHR-4). | |||||
|
CO2_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 37) | NC score | 0.460684 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
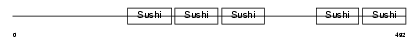
FHR5_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 38) | NC score | 0.739543 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BXR6 | Gene names | CFHL5, FHR5 | |||
|
Domain Architecture |
|
|||||
| Description | Complement factor H-related protein 5 precursor (FHR-5). | |||||
|
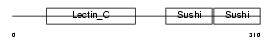
LYAM1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 39) | NC score | 0.517823 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
CO7_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 40) | NC score | 0.391413 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
SUSD4_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 41) | NC score | 0.722002 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
GABR1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 42) | NC score | 0.354203 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 43) | NC score | 0.352709 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
SUSD4_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 44) | NC score | 0.720975 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
SRPX_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 45) | NC score | 0.681148 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9R0M3, Q9R0M2 | Gene names | Srpx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi-repeat-containing protein SRPX precursor (DRS protein). | |||||
|
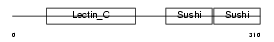
LYAM1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 46) | NC score | 0.523151 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
SRPX_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 47) | NC score | 0.665422 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P78539, Q99652, Q99913 | Gene names | SRPX, ETX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sushi repeat-containing protein SRPX precursor. | |||||
|
C1S_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 48) | NC score | 0.224123 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
MASP2_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 49) | NC score | 0.233777 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MASP1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 50) | NC score | 0.201999 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
SUSD1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 51) | NC score | 0.304242 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
CO2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 52) | NC score | 0.387636 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
HPT_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 53) | NC score | 0.108972 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P00738, P00737, Q2PP15, Q3B7J0, Q6LBY9 | Gene names | HP | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
SUSD2_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 54) | NC score | 0.442503 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
MASP1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 55) | NC score | 0.196516 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
CO6_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 56) | NC score | 0.326132 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
FHR3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 57) | NC score | 0.653503 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q02985, Q9UJ16 | Gene names | CFHL3, FHR3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 3 precursor (FHR-3) (H factor-like protein 3) (DOWN16). | |||||
|
SUSD2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 58) | NC score | 0.434807 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
MASP2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 59) | NC score | 0.213901 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
CS1B_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 60) | NC score | 0.199611 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
CS1A_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 61) | NC score | 0.199384 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
C1RA_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 62) | NC score | 0.186511 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 63) | NC score | 0.184967 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 64) | NC score | 0.150393 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
SUSD3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 65) | NC score | 0.367734 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D176, Q80XL5, Q9CYV1, Q9DA86 | Gene names | Susd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 66) | NC score | 0.143443 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
IL2RA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 67) | NC score | 0.163117 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P01589 | Gene names | IL2RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor alpha chain precursor (IL-2 receptor alpha subunit) (IL-2-RA) (IL2-RA) (p55) (TAC antigen) (CD25 antigen). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 68) | NC score | 0.124145 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 69) | NC score | 0.138070 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
ANRA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.008293 (rank : 166) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |
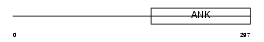
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
MGR6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.015906 (rank : 158) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
PERT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.176895 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
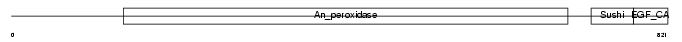
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.010884 (rank : 162) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
PERT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.155053 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P07202, P09934, P09935, Q8IUL0, Q8NF94, Q8NF95, Q8NF96, Q8NF97, Q8TCI9 | Gene names | TPO | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
ANRA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.007348 (rank : 168) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |
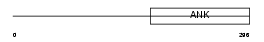
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.003967 (rank : 171) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SUSD3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.225584 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96L08, Q49AA6, Q6UXV7 | Gene names | SUSD3, UNQ9387/PRO34275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
TAF9B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.013662 (rank : 160) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBM6, Q9Y2S3 | Gene names | TAF9B, TAF9L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 9B (Transcription initiation factor TFIID subunit 9-like protein) (Transcription- associated factor TAFII31L) (Neuronal cell death-related protein 7) (DN-7). | |||||
|
ZNF30_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | -0.003542 (rank : 172) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17039, Q6N068 | Gene names | ZNF30, KOX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 30 (Zinc finger protein KOX28). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.122253 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.122855 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.010482 (rank : 163) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MGR6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.014974 (rank : 159) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.122090 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
DPEP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.010066 (rank : 165) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C255, Q3TAV9, Q3TBF7, Q3TBK3, Q3TBM9, Q7TQ53 | Gene names | Dpep2, Mbd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidase 2 precursor (EC 3.4.13.19) (Membrane-bound dipeptidase 2) (MBD-2). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.011962 (rank : 161) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ZNF23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | -0.003703 (rank : 173) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17027, Q96IT3, Q9UG42 | Gene names | ZNF23, KOX16, ZNF359 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 23 (Zinc finger protein 359) (Zinc finger protein KOX16). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.010440 (rank : 164) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.133413 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
SIA7A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.006439 (rank : 169) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
ZN432_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | -0.003773 (rank : 174) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94892 | Gene names | ZNF432, KIAA0798 | |||
|
Domain Architecture |
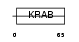
|
|||||
| Description | Zinc finger protein 432. | |||||
|
NFAC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.005375 (rank : 170) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.124251 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
ZN615_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | -0.003893 (rank : 175) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8J6 | Gene names | ZNF615 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 615. | |||||
|
K0247_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.222751 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92537 | Gene names | KIAA0247 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0247 precursor. | |||||
|
ZCH14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.008151 (rank : 167) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ZIC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | -0.004088 (rank : 176) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46684 | Gene names | Zic1, Zic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 1 (Zinc finger protein of the cerebellum 1). | |||||
|
ASGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.055427 (rank : 147) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
ASGR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.055964 (rank : 145) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
ASGR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.057875 (rank : 140) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
ATRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.063340 (rank : 127) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.061555 (rank : 133) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
BMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.124731 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.125008 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
C1R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.177606 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
C209A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.053932 (rank : 150) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZX1, Q3TAT9, Q91ZW5 | Gene names | Cd209a, Cire | |||
|
Domain Architecture |
|
|||||
| Description | CD209 antigen-like protein A (Dendritic cell-specific ICAM-3-grabbing nonintegrin) (DC-SIGN). | |||||
|
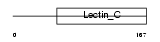
C209C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.054442 (rank : 149) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |
|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
C209E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.054684 (rank : 148) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZW7 | Gene names | Cd209e | |||
|
Domain Architecture |
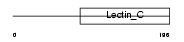
|
|||||
| Description | CD209 antigen-like protein E (DC-SIGN-related protein 4) (DC-SIGNR4). | |||||
|
CLC10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.059249 (rank : 137) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
CLC4A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.056328 (rank : 144) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QZ15, Q4VA33 | Gene names | Clec4a, Clec4a2, Clecsf6, Dcir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor). | |||||
|
CLC4C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.051150 (rank : 156) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WTT0, Q3T1C3, Q6UXS8, Q8WXX8 | Gene names | CLEC4C, BDCA2, CLECSF11, CLECSF7, DLEC, HECL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member C (C-type lectin superfamily member 7) (Blood dendritic cell antigen 2 protein) (BDCA-2) (Dendritic lectin) (CD303 antigen). | |||||
|
CLC4E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052754 (rank : 154) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULY5 | Gene names | CLEC4E, CLECSF9, MINCLE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.057116 (rank : 141) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.055506 (rank : 146) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
CLC4K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.065479 (rank : 122) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC4K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.064157 (rank : 125) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.107756 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.099367 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.102283 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.095634 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
DCBD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.067199 (rank : 121) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.087679 (rank : 107) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.073712 (rank : 116) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
DCBD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.067549 (rank : 120) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |
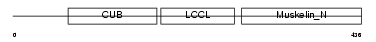
|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DMBT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.071928 (rank : 117) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.087594 (rank : 108) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
ENTK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.062402 (rank : 131) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
ENTK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.064892 (rank : 124) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
FA10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.060173 (rank : 134) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P00742, Q14340 | Gene names | F10 | |||
|
Domain Architecture |
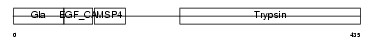
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) (Stuart- Prower factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
FA10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.059553 (rank : 136) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |
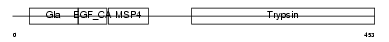
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
FA7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.050981 (rank : 157) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08709, Q14339, Q5JVF2 | Gene names | F7 | |||
|
Domain Architecture |
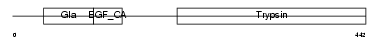
|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) (SPCA) (Proconvertin) (Eptacog alfa) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
FA9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052401 (rank : 155) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P00740 | Gene names | F9 | |||
|
Domain Architecture |
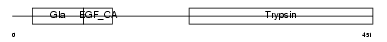
|
|||||
| Description | Coagulation factor IX precursor (EC 3.4.21.22) (Christmas factor) (Plasma thromboplastin component) (PTC) [Contains: Coagulation factor IXa light chain; Coagulation factor IXa heavy chain]. | |||||
|
FCER2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.058800 (rank : 138) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
GABR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.058183 (rank : 139) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
GP126_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.063002 (rank : 128) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
HPTR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.068192 (rank : 119) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00739, Q7LE20, Q92658, Q92659, Q9ULB0 | Gene names | HPR | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin-related protein precursor. | |||||
|
HPT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.071194 (rank : 118) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61646 | Gene names | Hp | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
I15RA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.062726 (rank : 129) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13261, Q6B0J2, Q7LDR4, Q7Z609 | Gene names | IL15RA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-15 receptor alpha chain precursor (IL-15R-alpha) (IL- 15RA). | |||||
|
I15RA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.091741 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60819, Q80Z90, Q80Z91, Q80Z92, Q8R5E4 | Gene names | Il15ra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-15 receptor alpha chain precursor (IL-15R-alpha) (IL- 15RA). | |||||
|
IL2RA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.053813 (rank : 152) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P01590, Q61731 | Gene names | Il2ra, Il2r | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor alpha chain precursor (IL-2 receptor alpha subunit) (IL-2-RA) (IL2-RA) (p55) (CD25 antigen). | |||||
|
KREM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.083520 (rank : 113) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96MU8, Q5TIB9, Q6P3X6, Q9BY70, Q9UGS5, Q9UGU1 | Gene names | KREMEN1, KREMEN, KRM1 | |||
|
Domain Architecture |
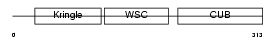
|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
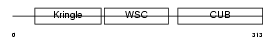
KREM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.086184 (rank : 111) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |
|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
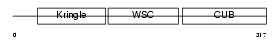
KREM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.082843 (rank : 114) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NCW0, Q8N2J4, Q8NCW1, Q96GL8, Q9BTP9 | Gene names | KREMEN2, KRM2 | |||
|
Domain Architecture |
|
|||||
| Description | Kremen protein 2 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 2) (Dickkopf receptor 2). | |||||
|
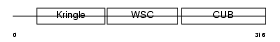
KREM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.075630 (rank : 115) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1S7 | Gene names | Kremen2 | |||
|
Domain Architecture |
|
|||||
| Description | Kremen protein 2 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 2) (Dickkopf receptor 2). | |||||
|
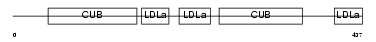
LRP12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.090892 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
LRP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.090658 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
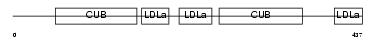
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
LRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.065231 (rank : 123) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
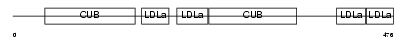
Domain Architecture |
|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
MFRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.105296 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.112141 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
MMGL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.056375 (rank : 143) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
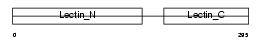
Domain Architecture |
|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
MRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.053201 (rank : 153) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
NETO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.090543 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.090219 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.087571 (rank : 109) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.086903 (rank : 110) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.086131 (rank : 112) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
NRP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.088447 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
NRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.096397 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
NRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.097696 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.121597 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.112894 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.109915 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PDGFD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.097052 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
PDGFD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.093576 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
PROC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.053872 (rank : 151) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04070, Q15189, Q15190, Q16001 | Gene names | PROC | |||
|
Domain Architecture |
|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
PROC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.056982 (rank : 142) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |
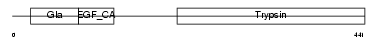
|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
PROZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.059962 (rank : 135) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |
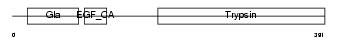
|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
PROZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.062353 (rank : 132) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CQW3 | Gene names | Proz | |||
|
Domain Architecture |
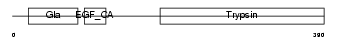
|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
ST14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.062446 (rank : 130) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
ST14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.063556 (rank : 126) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
TLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.125193 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
TLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.126866 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.125700 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.128544 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.108892 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.109854 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
DAF_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.0341e-181 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
DAF2_MOUSE
|
||||||
| NC score | 0.974351 (rank : 2) | θ value | 2.81514e-94 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q61476 | Gene names | Cd55b, Daf2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor transmembrane isoform precursor (DAF-TM) (CD55 antigen). | |||||
|
DAF1_MOUSE
|
||||||
| NC score | 0.965322 (rank : 3) | θ value | 2.37764e-101 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
CRRY_MOUSE
|
||||||
| NC score | 0.939843 (rank : 4) | θ value | 4.88048e-38 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q64735, Q3TV30, Q58E68, Q61447, Q61449, Q8CF59, Q8K328 | Gene names | Crry, Cry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement regulatory protein Crry precursor (Protein p65). | |||||
|
ZP3R_MOUSE
|
||||||
| NC score | 0.927491 (rank : 5) | θ value | 8.61488e-35 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q60736 | Gene names | Zp3r, Sp56 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 receptor precursor (Sperm fertilization protein 56) (sp56). | |||||
|
C4BP_MOUSE
|
||||||
| NC score | 0.924905 (rank : 6) | θ value | 3.74554e-30 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P08607 | Gene names | C4bpa, C4bp | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein precursor (C4bp). | |||||
|
C4BP_HUMAN
|
||||||
| NC score | 0.924231 (rank : 7) | θ value | 1.85459e-37 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P04003 | Gene names | C4BPA, C4BP | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein alpha chain precursor (C4bp) (Proline-rich protein) (PRP). | |||||
|
CR1_HUMAN
|
||||||
| NC score | 0.910123 (rank : 8) | θ value | 1.2411e-41 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P17927, Q16745, Q9UQV2 | Gene names | CR1, C3BR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 1 precursor (C3b/C4b receptor) (CD35 antigen). | |||||
|
MCP_HUMAN
|
||||||
| NC score | 0.899960 (rank : 9) | θ value | 4.43474e-23 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P15529, Q15429, Q53GV9, Q5HY94, Q5VWS6, Q5VWS7, Q5VWS8, Q5VWS9, Q5VWT0, Q5VWT1, Q5VWT2, Q6N0A1, Q7Z3R5, Q9NNW2, Q9NNW3, Q9NNW4 | Gene names | CD46, MCP | |||
|
Domain Architecture |

|
|||||
| Description | Membrane cofactor protein precursor (Trophoblast leukocyte common antigen) (TLX) (CD46 antigen). | |||||
|
CR2_MOUSE
|
||||||
| NC score | 0.892569 (rank : 10) | θ value | 1.4233e-29 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P19070 | Gene names | Cr2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (CD21 antigen). | |||||
|
CR2_HUMAN
|
||||||
| NC score | 0.889103 (rank : 11) | θ value | 1.12775e-26 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P20023, Q13866, Q14212 | Gene names | CR2, C3DR | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (Epstein-Barr virus receptor) (EBV receptor) (CD21 antigen). | |||||
|
MCP_MOUSE
|
||||||
| NC score | 0.885526 (rank : 12) | θ value | 5.60996e-18 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88174, Q9R0R9 | Gene names | Cd46, Mcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane cofactor protein precursor (CD46 antigen). | |||||
|
APOH_MOUSE
|
||||||
| NC score | 0.861599 (rank : 13) | θ value | 9.90251e-15 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q01339 | Gene names | Apoh, B2gp1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor). | |||||
|
APOH_HUMAN
|
||||||
| NC score | 0.854769 (rank : 14) | θ value | 1.38178e-16 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P02749, Q9UCN7 | Gene names | APOH, B2G1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-glycoprotein 1 precursor (Beta-2-glycoprotein I) (Apolipoprotein H) (Apo-H) (B2GPI) (Beta(2)GPI) (Activated protein C- binding protein) (APC inhibitor) (Anticardiolipin cofactor). | |||||
|
C4BB_HUMAN
|
||||||
| NC score | 0.846104 (rank : 15) | θ value | 2.28291e-11 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P20851 | Gene names | C4BPB | |||
|
Domain Architecture |

|
|||||
| Description | C4b-binding protein beta chain precursor. | |||||
|
CFAH_MOUSE
|
||||||
| NC score | 0.815744 (rank : 16) | θ value | 4.58923e-20 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P06909 | Gene names | Cfh, Hf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (Protein beta-1-H). | |||||
|
F13B_MOUSE
|
||||||
| NC score | 0.806945 (rank : 17) | θ value | 3.18553e-13 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
CFAH_HUMAN
|
||||||
| NC score | 0.803044 (rank : 18) | θ value | 4.44505e-15 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P08603, P78435, Q14570, Q38G77, Q5TFM3, Q8N708, Q9NU86 | Gene names | CFH, HF, HF1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H precursor (H factor 1). | |||||
|
F13B_HUMAN
|
||||||
| NC score | 0.796676 (rank : 19) | θ value | 2.0648e-12 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P05160 | Gene names | F13B | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain) (Fibrin- stabilizing factor B subunit). | |||||
|
FHR5_HUMAN
|
||||||
| NC score | 0.739543 (rank : 20) | θ value | 1.17247e-07 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BXR6 | Gene names | CFHL5, FHR5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 5 precursor (FHR-5). | |||||
|
SUSD4_HUMAN
|
||||||
| NC score | 0.722002 (rank : 21) | θ value | 1.99992e-07 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5VX71, Q6UX62, Q9BSR0, Q9NWG0 | Gene names | SUSD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
SUSD4_MOUSE
|
||||||
| NC score | 0.720975 (rank : 22) | θ value | 7.59969e-07 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BH32, Q8VC43 | Gene names | Susd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 4 precursor. | |||||
|
LYAM3_HUMAN
|
||||||
| NC score | 0.715699 (rank : 23) | θ value | 8.95645e-16 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P16109, Q8IVD1 | Gene names | SELP, GMRP | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
FHR1_HUMAN
|
||||||
| NC score | 0.715218 (rank : 24) | θ value | 2.69671e-12 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q03591, Q9UJ17 | Gene names | CFHL1, CFHL, FHR1, HFL1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 1 precursor (FHR-1) (H factor-like protein 1) (H-factor-like 1) (H36). | |||||
|
LYAM3_MOUSE
|
||||||
| NC score | 0.711742 (rank : 25) | θ value | 1.13037e-18 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q01102 | Gene names | Selp, Grmp | |||
|
Domain Architecture |

|
|||||
| Description | P-selectin precursor (Granule membrane protein 140) (GMP-140) (PADGEM) (Leukocyte-endothelial cell adhesion molecule 3) (LECAM3) (CD62P antigen). | |||||
|
LYAM2_HUMAN
|
||||||
| NC score | 0.697776 (rank : 26) | θ value | 2.7842e-17 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P16581, P16111 | Gene names | SELE, ELAM1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
CSMD3_HUMAN
|
||||||
| NC score | 0.695266 (rank : 27) | θ value | 1.99067e-23 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.689352 (rank : 28) | θ value | 3.39556e-23 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
LYAM2_MOUSE
|
||||||
| NC score | 0.685390 (rank : 29) | θ value | 2.88119e-14 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q00690 | Gene names | Sele, Elam-1 | |||
|
Domain Architecture |

|
|||||
| Description | E-selectin precursor (Endothelial leukocyte adhesion molecule 1) (ELAM-1) (Leukocyte-endothelial cell adhesion molecule 2) (LECAM2) (CD62E antigen). | |||||
|
CSMD1_MOUSE
|
||||||
| NC score | 0.683805 (rank : 30) | θ value | 1.09232e-21 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
SRPX_MOUSE
|
||||||
| NC score | 0.681148 (rank : 31) | θ value | 9.92553e-07 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9R0M3, Q9R0M2 | Gene names | Srpx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi-repeat-containing protein SRPX precursor (DRS protein). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.678097 (rank : 32) | θ value | 1.38178e-16 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.673297 (rank : 33) | θ value | 5.07402e-19 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
SRPX_HUMAN
|
||||||
| NC score | 0.665422 (rank : 34) | θ value | 2.21117e-06 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P78539, Q99652, Q99913 | Gene names | SRPX, ETX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sushi repeat-containing protein SRPX precursor. | |||||
|
FHR2_HUMAN
|
||||||
| NC score | 0.661551 (rank : 35) | θ value | 4.76016e-09 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P36980, Q14310 | Gene names | CFHL2, FHR2, HFL3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 2 precursor (FHR-2) (H factor-like protein 2) (H factor-like 3) (DDESK59). | |||||
|
FHR4_HUMAN
|
||||||
| NC score | 0.658303 (rank : 36) | θ value | 8.11959e-09 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q92496, Q9UJY6 | Gene names | CFHL4, FHR4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 4 precursor (FHR-4). | |||||
|
FHR3_HUMAN
|
||||||
| NC score | 0.653503 (rank : 37) | θ value | 0.00390308 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q02985, Q9UJ16 | Gene names | CFHL3, FHR3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor H-related protein 3 precursor (FHR-3) (H factor-like protein 3) (DOWN16). | |||||
|
PAPPA_HUMAN
|
||||||
| NC score | 0.607923 (rank : 38) | θ value | 4.76016e-09 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PAPPA_MOUSE
|
||||||
| NC score | 0.603424 (rank : 39) | θ value | 1.33837e-11 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PAP2_HUMAN
|
||||||
| NC score | 0.555361 (rank : 40) | θ value | 1.58096e-12 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
LYAM1_MOUSE
|
||||||
| NC score | 0.523151 (rank : 41) | θ value | 2.21117e-06 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
LYAM1_HUMAN
|
||||||
| NC score | 0.517823 (rank : 42) | θ value | 1.53129e-07 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
CFAB_MOUSE
|
||||||
| NC score | 0.466238 (rank : 43) | θ value | 3.64472e-09 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CFAB_HUMAN
|
||||||
| NC score | 0.463268 (rank : 44) | θ value | 6.64225e-11 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P00751, O15006, Q29944, Q96HX6, Q9BTF5, Q9BX92 | Gene names | CFB, BF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) (Properdin factor B) (Glycine-rich beta glycoprotein) (GBG) (PBF2) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CO2_HUMAN
|
||||||
| NC score | 0.460684 (rank : 45) | θ value | 2.36244e-08 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
SUSD2_MOUSE
|
||||||
| NC score | 0.442503 (rank : 46) | θ value | 0.000786445 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
SUSD2_HUMAN
|
||||||
| NC score | 0.434807 (rank : 47) | θ value | 0.00509761 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
CO7_HUMAN
|
||||||
| NC score | 0.391413 (rank : 48) | θ value | 1.99992e-07 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
CO2_MOUSE
|
||||||
| NC score | 0.387636 (rank : 49) | θ value | 0.000461057 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
SUSD3_MOUSE
|
||||||
| NC score | 0.367734 (rank : 50) | θ value | 0.0563607 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D176, Q80XL5, Q9CYV1, Q9DA86 | Gene names | Susd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
GABR1_HUMAN
|
||||||
| NC score | 0.354203 (rank : 51) | θ value | 4.45536e-07 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UBS5, O95375, O95468, O95975, O96022, Q86W60, Q9UQQ0 | Gene names | GABBR1, GPRC3A | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.352709 (rank : 52) | θ value | 4.45536e-07 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
CO6_HUMAN
|
||||||
| NC score | 0.326132 (rank : 53) | θ value | 0.00298849 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
SUSD1_HUMAN
|
||||||
| NC score | 0.304242 (rank : 54) | θ value | 0.000270298 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
MASP2_MOUSE
|
||||||
| NC score | 0.233777 (rank : 55) | θ value | 4.1701e-05 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
SUSD3_HUMAN
|
||||||
| NC score | 0.225584 (rank : 56) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96L08, Q49AA6, Q6UXV7 | Gene names | SUSD3, UNQ9387/PRO34275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 3. | |||||
|
C1S_HUMAN
|
||||||
| NC score | 0.224123 (rank : 57) | θ value | 2.44474e-05 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
K0247_HUMAN
|
||||||
| NC score | 0.222751 (rank : 58) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92537 | Gene names | KIAA0247 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0247 precursor. | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.213901 (rank : 59) | θ value | 0.00869519 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.201999 (rank : 60) | θ value | 0.000121331 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
CS1B_MOUSE
|
||||||
| NC score | 0.199611 (rank : 61) | θ value | 0.0113563 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
CS1A_MOUSE
|
||||||
| NC score | 0.199384 (rank : 62) | θ value | 0.0148317 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| NC score | 0.196516 (rank : 63) | θ value | 0.00102713 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
C1RA_MOUSE
|
||||||
| NC score | 0.186511 (rank : 64) | θ value | 0.0193708 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| NC score | 0.184967 (rank : 65) | θ value | 0.0330416 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
C1R_HUMAN
|
||||||
| NC score | 0.177606 (rank : 66) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
PERT_MOUSE
|
||||||
| NC score | 0.176895 (rank : 67) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
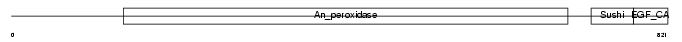
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
IL2RA_HUMAN
|
||||||
| NC score | 0.163117 (rank : 68) | θ value | 0.279714 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P01589 | Gene names | IL2RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor alpha chain precursor (IL-2 receptor alpha subunit) (IL-2-RA) (IL2-RA) (p55) (TAC antigen) (CD25 antigen). | |||||
|
PERT_HUMAN
|
||||||
| NC score | 0.155053 (rank : 69) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P07202, P09934, P09935, Q8IUL0, Q8NF94, Q8NF95, Q8NF96, Q8NF97, Q8TCI9 | Gene names | TPO | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.150393 (rank : 70) | θ value | 0.0431538 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.143443 (rank : 71) | θ value | 0.163984 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.138070 (rank : 72) | θ value | 0.47712 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.133413 (rank : 73) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
TLL2_MOUSE
|
||||||
| NC score | 0.128544 (rank : 74) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL1_MOUSE
|
||||||
| NC score | 0.126866 (rank : 75) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62381, Q3UTT9, Q8BNP5 | Gene names | Tll1, Tll | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-) (mTll). | |||||
|
TLL2_HUMAN
|
||||||
| NC score | 0.125700 (rank : 76) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL1_HUMAN
|
||||||
| NC score | 0.125193 (rank : 77) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
BMP1_MOUSE
|
||||||
| NC score | 0.125008 (rank : 78) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P98063 | Gene names | Bmp1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
BMP1_HUMAN
|
||||||
| NC score | 0.124731 (rank : 79) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P13497, Q13292, Q13872, Q14874, Q99421, Q99422, Q99423, Q9UL38 | Gene names | BMP1, PCOLC | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 1 precursor (EC 3.4.24.19) (BMP-1) (Procollagen C-proteinase) (PCP) (Mammalian tolloid protein) (mTld). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.124251 (rank : 80) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.124145 (rank : 81) | θ value | 0.279714 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.122855 (rank : 82) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.122253 (rank : 83) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.122090 (rank : 84) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.121597 (rank : 85) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
PCOC2_HUMAN
|
||||||
| NC score | 0.112894 (rank : 86) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.112141 (rank : 87) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
PCOC2_MOUSE
|
||||||
| NC score | 0.109915 (rank : 88) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.109854 (rank : 89) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
HPT_HUMAN
|
||||||
| NC score | 0.108972 (rank : 90) | θ value | 0.000602161 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P00738, P00737, Q2PP15, Q3B7J0, Q6LBY9 | Gene names | HP | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.108892 (rank : 91) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.107756 (rank : 92) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
MFRP_HUMAN
|
||||||
| NC score | 0.105296 (rank : 93) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BY79, Q335M3, Q96DQ9 | Gene names | MFRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.102283 (rank : 94) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.099367 (rank : 95) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
NRP2_MOUSE
|
||||||
| NC score | 0.097696 (rank : 96) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
PDGFD_HUMAN
|
||||||
| NC score | 0.097052 (rank : 97) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9GZP0, Q9BWV5 | Gene names | PDGFD, IEGF, SCDGFB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor D precursor (PDGF D) (Iris-expressed growth factor) (Spinal cord-derived growth factor B) (SCDGF-B). | |||||
|
NRP2_HUMAN
|
||||||
| NC score | 0.096397 (rank : 98) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.095634 (rank : 99) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
PDGFD_MOUSE
|
||||||
| NC score | 0.093576 (rank : 100) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
I15RA_MOUSE
|
||||||
| NC score | 0.091741 (rank : 101) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60819, Q80Z90, Q80Z91, Q80Z92, Q8R5E4 | Gene names | Il15ra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-15 receptor alpha chain precursor (IL-15R-alpha) (IL- 15RA). | |||||
|
LRP12_HUMAN
|
||||||
| NC score | 0.090892 (rank : 102) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
LRP12_MOUSE
|
||||||
| NC score | 0.090658 (rank : 103) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BUJ9, Q8BWM9 | Gene names | Lrp12 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor. | |||||
|
NETO1_HUMAN
|
||||||
| NC score | 0.090543 (rank : 104) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDF5, Q86W85, Q8ND78, Q8TDF4 | Gene names | NETO1, BTCL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NETO1_MOUSE
|
||||||
| NC score | 0.090219 (rank : 105) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4I7, Q80X39, Q8C4S3, Q8CCM2 | Gene names | Neto1, Btcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 1 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 1). | |||||
|
NRP1_MOUSE
|
||||||
| NC score | 0.088447 (rank : 106) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97333 | Gene names | Nrp1, Nrp | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (A5 protein). | |||||
|
DCBD1_MOUSE
|
||||||
| NC score | 0.087679 (rank : 107) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.087594 (rank : 108) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
NETO2_HUMAN
|
||||||
| NC score | 0.087571 (rank : 109) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NC67, Q7Z381, Q8ND51, Q96SP4, Q9NVY8 | Gene names | NETO2, BTCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
NETO2_MOUSE
|
||||||
| NC score | 0.086903 (rank : 110) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BNJ6, Q5VM49, Q8C4Q8 | Gene names | Neto2, Btcl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuropilin and tolloid-like protein 2 precursor (Brain-specific transmembrane protein containing 2 CUB and 1 LDL-receptor class A domains protein 2). | |||||
|
KREM1_MOUSE
|
||||||
| NC score | 0.086184 (rank : 111) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
NRP1_HUMAN
|
||||||
| NC score | 0.086131 (rank : 112) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14786, O60461, Q96IH5 | Gene names | NRP1, NRP, VEGF165R | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-1 precursor (Vascular endothelial cell growth factor 165 receptor) (CD304 antigen). | |||||
|
KREM1_HUMAN
|
||||||
| NC score | 0.083520 (rank : 113) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96MU8, Q5TIB9, Q6P3X6, Q9BY70, Q9UGS5, Q9UGU1 | Gene names | KREMEN1, KREMEN, KRM1 | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
KREM2_HUMAN
|
||||||
| NC score | 0.082843 (rank : 114) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NCW0, Q8N2J4, Q8NCW1, Q96GL8, Q9BTP9 | Gene names | KREMEN2, KRM2 | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 2 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 2) (Dickkopf receptor 2). | |||||
|
KREM2_MOUSE
|
||||||
| NC score | 0.075630 (rank : 115) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K1S7 | Gene names | Kremen2 | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 2 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 2) (Dickkopf receptor 2). | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.073712 (rank : 116) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
DMBT1_HUMAN
|
||||||
| NC score | 0.071928 (rank : 117) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
HPT_MOUSE
|
||||||
| NC score | 0.071194 (rank : 118) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61646 | Gene names | Hp | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
HPTR_HUMAN
|
||||||
| NC score | 0.068192 (rank : 119) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00739, Q7LE20, Q92658, Q92659, Q9ULB0 | Gene names | HPR | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin-related protein precursor. | |||||
|
DCBD2_MOUSE
|
||||||
| NC score | 0.067549 (rank : 120) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD1_HUMAN
|
||||||
| NC score | 0.067199 (rank : 121) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
CLC4K_HUMAN
|
||||||
| NC score | 0.065479 (rank : 122) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
LRP3_HUMAN
|
||||||
| NC score | 0.065231 (rank : 123) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
ENTK_MOUSE
|
||||||
| NC score | 0.064892 (rank : 124) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
CLC4K_MOUSE
|
||||||
| NC score | 0.064157 (rank : 125) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
ST14_MOUSE
|
||||||
| NC score | 0.063556 (rank : 126) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.063340 (rank : 127) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
GP126_HUMAN
|
||||||
| NC score | 0.063002 (rank : 128) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
I15RA_HUMAN
|
||||||
| NC score | 0.062726 (rank : 129) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13261, Q6B0J2, Q7LDR4, Q7Z609 | Gene names | IL15RA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-15 receptor alpha chain precursor (IL-15R-alpha) (IL- 15RA). | |||||
|
ST14_HUMAN
|
||||||
| NC score | 0.062446 (rank : 130) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
ENTK_HUMAN
|
||||||
| NC score | 0.062402 (rank : 131) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
PROZ_MOUSE
|
||||||
| NC score | 0.062353 (rank : 132) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CQW3 | Gene names | Proz | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.061555 (rank : 133) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
FA10_HUMAN
|
||||||
| NC score | 0.060173 (rank : 134) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P00742, Q14340 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) (Stuart- Prower factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
PROZ_HUMAN
|
||||||
| NC score | 0.059962 (rank : 135) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
FA10_MOUSE
|
||||||
| NC score | 0.059553 (rank : 136) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
CLC10_HUMAN
|
||||||
| NC score | 0.059249 (rank : 137) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
FCER2_HUMAN
|
||||||
| NC score | 0.058800 (rank : 138) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
GABR2_HUMAN
|
||||||
| NC score | 0.058183 (rank : 139) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75899, O75974, O75975, Q5VXZ2, Q8WX04, Q9P1R2, Q9UNR1, Q9UNS9 | Gene names | GABBR2, GPR51, GPRC3B | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 2 precursor (GABA-B receptor 2) (GABA-B-R2) (Gb2) (GABABR2) (G-protein coupled receptor 51) (HG20). | |||||
|
ASGR2_MOUSE
|
||||||
| NC score | 0.057875 (rank : 140) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
CLC4E_MOUSE
|
||||||
| NC score | 0.057116 (rank : 141) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
PROC_MOUSE
|
||||||
| NC score | 0.056982 (rank : 142) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
MMGL_MOUSE
|
||||||
| NC score | 0.056375 (rank : 143) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
CLC4A_MOUSE
|
||||||
| NC score | 0.056328 (rank : 144) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QZ15, Q4VA33 | Gene names | Clec4a, Clec4a2, Clecsf6, Dcir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor). | |||||
|
ASGR2_HUMAN
|
||||||
| NC score | 0.055964 (rank : 145) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
CLC4G_MOUSE
|
||||||
| NC score | 0.055506 (rank : 146) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
ASGR1_MOUSE
|
||||||
| NC score | 0.055427 (rank : 147) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
C209E_MOUSE
|
||||||
| NC score | 0.054684 (rank : 148) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZW7 | Gene names | Cd209e | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein E (DC-SIGN-related protein 4) (DC-SIGNR4). | |||||
|
C209C_MOUSE
|
||||||
| NC score | 0.054442 (rank : 149) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
C209A_MOUSE
|
||||||
| NC score | 0.053932 (rank : 150) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91ZX1, Q3TAT9, Q91ZW5 | Gene names | Cd209a, Cire | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein A (Dendritic cell-specific ICAM-3-grabbing nonintegrin) (DC-SIGN). | |||||
|
PROC_HUMAN
|
||||||
| NC score | 0.053872 (rank : 151) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04070, Q15189, Q15190, Q16001 | Gene names | PROC | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
IL2RA_MOUSE
|
||||||
| NC score | 0.053813 (rank : 152) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P01590, Q61731 | Gene names | Il2ra, Il2r | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor alpha chain precursor (IL-2 receptor alpha subunit) (IL-2-RA) (IL2-RA) (p55) (CD25 antigen). | |||||
|
MRC1_HUMAN
|
||||||
| NC score | 0.053201 (rank : 153) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
CLC4E_HUMAN
|
||||||
| NC score | 0.052754 (rank : 154) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULY5 | Gene names | CLEC4E, CLECSF9, MINCLE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
FA9_HUMAN
|
||||||
| NC score | 0.052401 (rank : 155) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P00740 | Gene names | F9 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor IX precursor (EC 3.4.21.22) (Christmas factor) (Plasma thromboplastin component) (PTC) [Contains: Coagulation factor IXa light chain; Coagulation factor IXa heavy chain]. | |||||
|
CLC4C_HUMAN
|
||||||
| NC score | 0.051150 (rank : 156) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WTT0, Q3T1C3, Q6UXS8, Q8WXX8 | Gene names | CLEC4C, BDCA2, CLECSF11, CLECSF7, DLEC, HECL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member C (C-type lectin superfamily member 7) (Blood dendritic cell antigen 2 protein) (BDCA-2) (Dendritic lectin) (CD303 antigen). | |||||
|
FA7_HUMAN
|
||||||
| NC score | 0.050981 (rank : 157) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08709, Q14339, Q5JVF2 | Gene names | F7 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) (SPCA) (Proconvertin) (Eptacog alfa) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
MGR6_MOUSE
|
||||||
| NC score | 0.015906 (rank : 158) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR6_HUMAN
|
||||||
| NC score | 0.014974 (rank : 159) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
TAF9B_HUMAN
|
||||||
| NC score | 0.013662 (rank : 160) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBM6, Q9Y2S3 | Gene names | TAF9B, TAF9L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 9B (Transcription initiation factor TFIID subunit 9-like protein) (Transcription- associated factor TAFII31L) (Neuronal cell death-related protein 7) (DN-7). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.011962 (rank : 161) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.010884 (rank : 162) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.010482 (rank : 163) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.010440 (rank : 164) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
DPEP2_MOUSE
|
||||||
| NC score | 0.010066 (rank : 165) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C255, Q3TAV9, Q3TBF7, Q3TBK3, Q3TBM9, Q7TQ53 | Gene names | Dpep2, Mbd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidase 2 precursor (EC 3.4.13.19) (Membrane-bound dipeptidase 2) (MBD-2). | |||||
|
ANRA2_HUMAN
|
||||||
| NC score | 0.008293 (rank : 166) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ZCH14_MOUSE
|
||||||
| NC score | 0.008151 (rank : 167) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ANRA2_MOUSE
|
||||||
| NC score | 0.007348 (rank : 168) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.006439 (rank : 169) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
NFAC2_MOUSE
|
||||||
| NC score | 0.005375 (rank : 170) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.003967 (rank : 171) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ZNF30_HUMAN
|
||||||
| NC score | -0.003542 (rank : 172) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17039, Q6N068 | Gene names | ZNF30, KOX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 30 (Zinc finger protein KOX28). | |||||
|
ZNF23_HUMAN
|
||||||
| NC score | -0.003703 (rank : 173) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17027, Q96IT3, Q9UG42 | Gene names | ZNF23, KOX16, ZNF359 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 23 (Zinc finger protein 359) (Zinc finger protein KOX16). | |||||
|
ZN432_HUMAN
|
||||||
| NC score | -0.003773 (rank : 174) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94892 | Gene names | ZNF432, KIAA0798 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 432. | |||||
|
ZN615_HUMAN
|
||||||
| NC score | -0.003893 (rank : 175) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8J6 | Gene names | ZNF615 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 615. | |||||
|
ZIC1_MOUSE
|
||||||
| NC score | -0.004088 (rank : 176) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46684 | Gene names | Zic1, Zic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 1 (Zinc finger protein of the cerebellum 1). | |||||