Please be patient as the page loads
|
TNAP3_HUMAN
|
||||||
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TNAP3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
TNAP3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992183 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60769 | Gene names | Tnfaip3, Tnfip3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
OTU7B_HUMAN
|
||||||
| θ value | 2.18568e-46 (rank : 3) | NC score | 0.777660 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
OTU7A_HUMAN
|
||||||
| θ value | 1.12253e-42 (rank : 4) | NC score | 0.774302 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
OTU7A_MOUSE
|
||||||
| θ value | 1.12253e-42 (rank : 5) | NC score | 0.769134 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
ZRAN1_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 6) | NC score | 0.701019 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
VCIP1_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 7) | NC score | 0.326890 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JH7, Q504T4, Q86T93, Q86W01, Q8N3A9, Q9H5R8 | Gene names | VCPIP1, KIAA1850, VCIP135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
VCIP1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 8) | NC score | 0.327215 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CDG3, Q7TMU9, Q8BP90 | Gene names | Vcpip1, Vcip135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
ZFAN6_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.173710 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6FIF0, O95792, Q9BQF7, Q9GZY3 | Gene names | ZFAND6, AWP1, ZA20D3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 6 (Zinc finger A20 domain-containing protein 3) (Associated with PRK1 protein). | |||||
|
ZFAN6_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 10) | NC score | 0.163771 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DCH6, Q9ER79 | Gene names | Zfand6, Awp1, Za20d3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 6 (Zinc finger A20 domain-containing protein 3) (Associated with PRK1 protein). | |||||
|
ZN467_MOUSE
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.004312 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZL0, Q9JJ98 | Gene names | Znf467, Ezi, Zfp467 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 467 (Endothelial cell-derived zinc finger protein) (EZI). | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.016229 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
ZFAN5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.165196 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O76080 | Gene names | ZFAND5, ZA20D2, ZNF216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
ZFAN5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.163787 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88878 | Gene names | Zfand5, Za20d2, Zfp216, Znf216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
DAAM2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.026859 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.017934 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.022429 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.014753 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
DAAM2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.025579 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
PCX1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.028166 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
TRIM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.011243 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.017121 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
GPNMB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.029857 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.014644 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
ZNHI4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.030208 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
CN115_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.065795 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8Y1, Q9NVC7 | Gene names | C14orf115 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C14orf115. | |||||
|
IRX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.012875 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78414, Q7Z2F8, Q8N312 | Gene names | IRX1, IRXA1 | |||
|
Domain Architecture |
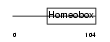
|
|||||
| Description | Iroquois-class homeodomain protein IRX-1 (Iroquois homeobox protein 1) (Homeodomain protein IRXA1). | |||||
|
RABX5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.077266 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJ41 | Gene names | RABGEF1, RABEX5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5) (Rabaptin-5-associated exchange factor for Rab5). | |||||
|
RABX5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.083121 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JM13, Q3UIW0 | Gene names | Rabgef1, Rabex5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.018030 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
K1949_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.020620 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
NET2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.018722 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1A3, Q9QY49, Q9WVA6 | Gene names | Ntn2l, Ntn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor (Netrin-3). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.015973 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.006638 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
NET2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.017858 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00634 | Gene names | NTN2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor. | |||||
|
PAD5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.013811 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68ED3, Q8C0K6 | Gene names | Papd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
PECI_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.012167 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.011898 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ADDA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.009629 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.014173 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
EGR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.002194 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.009818 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
TBCD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.008046 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYJ6, Q5DU23, Q66JU2, Q6P2M2, Q8BMH6, Q8BXM2 | Gene names | Tbc1d4, Kiaa0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
ABLM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.003840 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6H8Q1, Q6H8Q0, Q6NX73, Q8N3C5, Q8N9E9, Q8N9G2, Q96JL7 | Gene names | ABLIM2, KIAA1808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 2 (Actin-binding LIM protein family member 2) (abLIM-2). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.003435 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.004386 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CHGUT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.009125 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.008270 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DYDC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.017474 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IM9, Q5QP07, Q5QP11 | Gene names | DYDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 2. | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.017112 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
PAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.019710 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
SPAG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.009071 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPE6, O43648 | Gene names | SPAG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 4 protein (Outer dense fiber-associated protein SPAG4). | |||||
|
UBA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.014170 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.014160 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.009060 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ZN260_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.000725 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62513, Q3U170, Q8C2Q6, Q9R2B4 | Gene names | Znf260, Zfp260 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 260 (Zfp-260). | |||||
|
ZN646_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.002045 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
ZNF44_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.000766 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15621, P17018, Q9ULZ7 | Gene names | ZNF44, KOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 44 (Zinc finger protein KOX7) (Gonadotropin- inducible transcription repressor 2) (GIOT-2). | |||||
|
TNAP3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
TNAP3_MOUSE
|
||||||
| NC score | 0.992183 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60769 | Gene names | Tnfaip3, Tnfip3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
OTU7B_HUMAN
|
||||||
| NC score | 0.777660 (rank : 3) | θ value | 2.18568e-46 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
OTU7A_HUMAN
|
||||||
| NC score | 0.774302 (rank : 4) | θ value | 1.12253e-42 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
OTU7A_MOUSE
|
||||||
| NC score | 0.769134 (rank : 5) | θ value | 1.12253e-42 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
ZRAN1_HUMAN
|
||||||
| NC score | 0.701019 (rank : 6) | θ value | 8.3442e-30 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
VCIP1_MOUSE
|
||||||
| NC score | 0.327215 (rank : 7) | θ value | 3.08544e-08 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CDG3, Q7TMU9, Q8BP90 | Gene names | Vcpip1, Vcip135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
VCIP1_HUMAN
|
||||||
| NC score | 0.326890 (rank : 8) | θ value | 3.08544e-08 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JH7, Q504T4, Q86T93, Q86W01, Q8N3A9, Q9H5R8 | Gene names | VCPIP1, KIAA1850, VCIP135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
ZFAN6_HUMAN
|
||||||
| NC score | 0.173710 (rank : 9) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6FIF0, O95792, Q9BQF7, Q9GZY3 | Gene names | ZFAND6, AWP1, ZA20D3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 6 (Zinc finger A20 domain-containing protein 3) (Associated with PRK1 protein). | |||||
|
ZFAN5_HUMAN
|
||||||
| NC score | 0.165196 (rank : 10) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O76080 | Gene names | ZFAND5, ZA20D2, ZNF216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
ZFAN5_MOUSE
|
||||||
| NC score | 0.163787 (rank : 11) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88878 | Gene names | Zfand5, Za20d2, Zfp216, Znf216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
ZFAN6_MOUSE
|
||||||
| NC score | 0.163771 (rank : 12) | θ value | 0.0330416 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DCH6, Q9ER79 | Gene names | Zfand6, Awp1, Za20d3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 6 (Zinc finger A20 domain-containing protein 3) (Associated with PRK1 protein). | |||||
|
RABX5_MOUSE
|
||||||
| NC score | 0.083121 (rank : 13) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JM13, Q3UIW0 | Gene names | Rabgef1, Rabex5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5). | |||||
|
RABX5_HUMAN
|
||||||
| NC score | 0.077266 (rank : 14) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJ41 | Gene names | RABGEF1, RABEX5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5) (Rabaptin-5-associated exchange factor for Rab5). | |||||
|
CN115_HUMAN
|
||||||
| NC score | 0.065795 (rank : 15) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8Y1, Q9NVC7 | Gene names | C14orf115 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C14orf115. | |||||
|
ZNHI4_MOUSE
|
||||||
| NC score | 0.030208 (rank : 16) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
GPNMB_HUMAN
|
||||||
| NC score | 0.029857 (rank : 17) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
PCX1_HUMAN
|
||||||
| NC score | 0.028166 (rank : 18) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
DAAM2_MOUSE
|
||||||
| NC score | 0.026859 (rank : 19) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U19, Q810J5 | Gene names | Daam2, Kiaa0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
DAAM2_HUMAN
|
||||||
| NC score | 0.025579 (rank : 20) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.022429 (rank : 21) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
K1949_MOUSE
|
||||||
| NC score | 0.020620 (rank : 22) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BQ30, Q3UDS6, Q8CEY9, Q8R3D8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949 homolog. | |||||
|
PAP2_HUMAN
|
||||||
| NC score | 0.019710 (rank : 23) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXP8, Q96PH7, Q96PH8, Q9H4C9 | Gene names | PAPPA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-2 precursor (EC 3.4.24.-) (Pregnancy-associated plasma protein-A2) (PAPP-A2) (Pregnancy-associated plasma protein-E1) (PAPP- E). | |||||
|
NET2_MOUSE
|
||||||
| NC score | 0.018722 (rank : 24) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1A3, Q9QY49, Q9WVA6 | Gene names | Ntn2l, Ntn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor (Netrin-3). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.018030 (rank : 25) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.017934 (rank : 26) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
NET2_HUMAN
|
||||||
| NC score | 0.017858 (rank : 27) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00634 | Gene names | NTN2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor. | |||||
|
DYDC2_HUMAN
|
||||||
| NC score | 0.017474 (rank : 28) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IM9, Q5QP07, Q5QP11 | Gene names | DYDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DPY30 domain-containing protein 2. | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.017121 (rank : 29) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.017112 (rank : 30) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.016229 (rank : 31) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.015973 (rank : 32) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.014753 (rank : 33) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.014644 (rank : 34) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.014173 (rank : 35) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
UBA3_HUMAN
|
||||||
| NC score | 0.014170 (rank : 36) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TBC4, O76088, Q9NTU3 | Gene names | UBE1C, UBA3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
UBA3_MOUSE
|
||||||
| NC score | 0.014160 (rank : 37) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C878, O88598, Q9D6B7 | Gene names | Ube1c, Uba3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD8-activating enzyme E1 catalytic subunit (EC 6.3.2.-) (Ubiquitin- activating enzyme 3) (NEDD8-activating enzyme E1C) (Ubiquitin- activating enzyme E1C). | |||||
|
PAD5_MOUSE
|
||||||
| NC score | 0.013811 (rank : 38) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q68ED3, Q8C0K6 | Gene names | Papd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
IRX1_HUMAN
|
||||||
| NC score | 0.012875 (rank : 39) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78414, Q7Z2F8, Q8N312 | Gene names | IRX1, IRXA1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-1 (Iroquois homeobox protein 1) (Homeodomain protein IRXA1). | |||||
|
PECI_HUMAN
|
||||||
| NC score | 0.012167 (rank : 40) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.011898 (rank : 41) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TRIM1_HUMAN
|
||||||
| NC score | 0.011243 (rank : 42) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJV3, Q5JYF5, Q8WWK1, Q9UJR9 | Gene names | MID2, FXY2, RNF60, TRIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Midline-2 (Midline defect 2) (Tripartite motif-containing protein 1) (Midin-2) (RING finger protein 60). | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.009818 (rank : 43) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
ADDA_HUMAN
|
||||||
| NC score | 0.009629 (rank : 44) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
CHGUT_HUMAN
|
||||||
| NC score | 0.009125 (rank : 45) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E5, Q6P2I4, Q6UXD2 | Gene names | CSGLCAT, KIAA1402 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate glucuronyltransferase (EC 2.4.1.226) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase) (Chondroitin glucuronyltransferase II) (CSGlcA-T). | |||||
|
SPAG4_HUMAN
|
||||||
| NC score | 0.009071 (rank : 46) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPE6, O43648 | Gene names | SPAG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 4 protein (Outer dense fiber-associated protein SPAG4). | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.009060 (rank : 47) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.008270 (rank : 48) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
TBCD4_MOUSE
|
||||||
| NC score | 0.008046 (rank : 49) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BYJ6, Q5DU23, Q66JU2, Q6P2M2, Q8BMH6, Q8BXM2 | Gene names | Tbc1d4, Kiaa0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.006638 (rank : 50) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.004386 (rank : 51) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
ZN467_MOUSE
|
||||||
| NC score | 0.004312 (rank : 52) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZL0, Q9JJ98 | Gene names | Znf467, Ezi, Zfp467 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 467 (Endothelial cell-derived zinc finger protein) (EZI). | |||||
|
ABLM2_HUMAN
|
||||||
| NC score | 0.003840 (rank : 53) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6H8Q1, Q6H8Q0, Q6NX73, Q8N3C5, Q8N9E9, Q8N9G2, Q96JL7 | Gene names | ABLIM2, KIAA1808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 2 (Actin-binding LIM protein family member 2) (abLIM-2). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.003435 (rank : 54) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
EGR2_HUMAN
|
||||||
| NC score | 0.002194 (rank : 55) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
ZN646_HUMAN
|
||||||
| NC score | 0.002045 (rank : 56) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
ZNF44_HUMAN
|
||||||
| NC score | 0.000766 (rank : 57) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15621, P17018, Q9ULZ7 | Gene names | ZNF44, KOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 44 (Zinc finger protein KOX7) (Gonadotropin- inducible transcription repressor 2) (GIOT-2). | |||||
|
ZN260_MOUSE
|
||||||
| NC score | 0.000725 (rank : 58) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62513, Q3U170, Q8C2Q6, Q9R2B4 | Gene names | Znf260, Zfp260 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 260 (Zfp-260). | |||||