Please be patient as the page loads
|
ZRAN1_HUMAN
|
||||||
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZRAN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
TNAP3_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 2) | NC score | 0.707938 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60769 | Gene names | Tnfaip3, Tnfip3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
TNAP3_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 3) | NC score | 0.701019 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
OTU7B_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 4) | NC score | 0.679850 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
OTU7A_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 5) | NC score | 0.684976 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
OTU7A_MOUSE
|
||||||
| θ value | 1.62847e-25 (rank : 6) | NC score | 0.680726 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 7) | NC score | 0.196760 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 8) | NC score | 0.191855 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 9) | NC score | 0.319819 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
RNF31_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 10) | NC score | 0.231959 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |
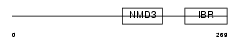
|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
RNF31_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 11) | NC score | 0.227783 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
NEIL3_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 12) | NC score | 0.150177 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 13) | NC score | 0.149190 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.131888 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
TAB2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.136584 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
YAF2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.190989 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IY57, Q99710 | Gene names | YAF2 | |||
|
Domain Architecture |
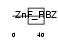
|
|||||
| Description | YY1-associated factor 2. | |||||
|
YAF2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.195626 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LW6, Q3UFD4, Q9DBE4 | Gene names | Yaf2 | |||
|
Domain Architecture |
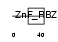
|
|||||
| Description | YY1-associated factor 2. | |||||
|
RYBP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.176625 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N488, Q9P2W5, Q9UMW4 | Gene names | RYBP, DEDAF, YEAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING1 and YY1-binding protein (Death effector domain-associated factor) (DED-associated factor) (YY1 and E4TF1-associated factor 1) (Apoptin-associating protein 1) (APAP-1). | |||||
|
RYBP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.173368 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CCI5, Q9WVK2 | Gene names | Rybp, Dedaf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING1 and YY1-binding protein (Death effector domain-associated factor) (DED-associated factor). | |||||
|
ZRAB2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.112546 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
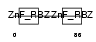
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.118010 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
R113B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.068405 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
SPN90_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.062974 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESJ4, Q68G72 | Gene names | Nckipsd, Spin90, Wasbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (N-WASP-interacting protein, 90kDa) (Wiskott-Aldrich syndrome protein-binding protein WISH) (N-WASP-binding protein). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.114005 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.009955 (rank : 66) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NPL4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.084066 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TAT6, Q8N3J1, Q9H8V2, Q9H964, Q9NWR5, Q9P229 | Gene names | NPLOC4, KIAA1499, NPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear protein localization protein 4 homolog (Protein NPL4). | |||||
|
NPL4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.084139 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60670 | Gene names | Nploc4, Kiaa1499, Npl4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear protein localization protein 4 homolog (Protein NPL4). | |||||
|
R113A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.060321 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
SPN90_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.057117 (rank : 49) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZQ3, Q9UGM8 | Gene names | NCKIPSD, AF3P21, SPIN90 | |||
|
Domain Architecture |
|
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (AF3p21) (Diaphanous protein-interacting protein) (Dia-interacting protein 1) (DIP-1). | |||||
|
EVPL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.013696 (rank : 65) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.024854 (rank : 56) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.025135 (rank : 55) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.008977 (rank : 68) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
ELOA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.023174 (rank : 57) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
SRGP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.022083 (rank : 58) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
UDA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.007267 (rank : 69) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4X1 | Gene names | UGT2A1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 2A1 precursor (EC 2.4.1.17). | |||||
|
MUG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.014572 (rank : 64) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28665 | Gene names | Mug1, Mug-1 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-1 precursor (MuG1). | |||||
|
MUG2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.014962 (rank : 63) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28666 | Gene names | Mug2, Mug-2 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-2 precursor (MuG2). | |||||
|
CDKL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | -0.001513 (rank : 70) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BLF2, Q5M6W2, Q8BKR2, Q8BL49, Q8BVE0, Q8K134 | Gene names | Cdkl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 3 (EC 2.7.11.22). | |||||
|
EWS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.016685 (rank : 60) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.016256 (rank : 62) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
GASP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.009352 (rank : 67) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.016282 (rank : 61) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.017676 (rank : 59) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.052137 (rank : 54) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPIA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.066256 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P62937, P05092, Q6IBU5, Q96IX3, Q9BRU4, Q9BTY9, Q9UC61 | Gene names | PPIA, CYPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein). | |||||
|
PPIA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.066306 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17742, Q9CWJ5, Q9R137 | Gene names | Ppia | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein) (SP18). | |||||
|
PPIB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.058811 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23284, Q9BVK5 | Gene names | PPIB, CYPB | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.058735 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24369 | Gene names | Ppib | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.058701 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P45877 | Gene names | PPIC, CYPC | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPIC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.058329 (rank : 48) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30412 | Gene names | Ppic, Cypc | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPID_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.052684 (rank : 51) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPID_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.053068 (rank : 50) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
PPIE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.052593 (rank : 53) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.052639 (rank : 52) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.065896 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30405, Q5W131 | Gene names | PPIF, CYP3 | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPIF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.066958 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KR7 | Gene names | Ppif | |||
|
Domain Architecture |
|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPIH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.059106 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43447 | Gene names | PPIH, CYP20, CYPH | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H) (U-snRNP-associated cyclophilin SnuCyp-20) (USA-CYP) (Small nuclear ribonucleoprotein particle-specific cyclophilin H) (CypH). | |||||
|
PPIH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.059617 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D868, Q9CQU7 | Gene names | Ppih | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H). | |||||
|
RANG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.110245 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43487 | Gene names | RANBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RanBP1). | |||||
|
RANG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.111029 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34022, P34023, Q3TL81, Q9D894, Q9DCA3 | Gene names | Ranbp1, Htf9-a, Htf9a | |||
|
Domain Architecture |
|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RANBP1) (HpaII tiny fragments locus 9a protein). | |||||
|
RBP22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.118589 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RBP26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.118302 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
RGPD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.118380 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
VCIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.111951 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JH7, Q504T4, Q86T93, Q86W01, Q8N3A9, Q9H5R8 | Gene names | VCPIP1, KIAA1850, VCIP135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
VCIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.112080 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CDG3, Q7TMU9, Q8BP90 | Gene names | Vcpip1, Vcip135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
ZFAN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.065882 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O76080 | Gene names | ZFAND5, ZA20D2, ZNF216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
ZFAN5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.065233 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88878 | Gene names | Zfand5, Za20d2, Zfp216, Znf216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
ZFAN6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.067501 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6FIF0, O95792, Q9BQF7, Q9GZY3 | Gene names | ZFAND6, AWP1, ZA20D3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 6 (Zinc finger A20 domain-containing protein 3) (Associated with PRK1 protein). | |||||
|
ZFAN6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.062136 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCH6, Q9ER79 | Gene names | Zfand6, Awp1, Za20d3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 6 (Zinc finger A20 domain-containing protein 3) (Associated with PRK1 protein). | |||||
|
ZRAN1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
TNAP3_MOUSE
|
||||||
| NC score | 0.707938 (rank : 2) | θ value | 4.89182e-30 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60769 | Gene names | Tnfaip3, Tnfip3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
TNAP3_HUMAN
|
||||||
| NC score | 0.701019 (rank : 3) | θ value | 8.3442e-30 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
OTU7A_HUMAN
|
||||||
| NC score | 0.684976 (rank : 4) | θ value | 1.24688e-25 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
OTU7A_MOUSE
|
||||||
| NC score | 0.680726 (rank : 5) | θ value | 1.62847e-25 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
OTU7B_HUMAN
|
||||||
| NC score | 0.679850 (rank : 6) | θ value | 3.87602e-27 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6GQQ9, Q8WWA7, Q9NQ53, Q9UFF4 | Gene names | OTUD7B, ZA20D1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7B (EC 3.-.-.-) (Zinc finger protein Cezanne) (Zinc finger A20 domain-containing protein 1) (Cellular zinc finger anti-NF-kappa B protein). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.319819 (rank : 7) | θ value | 3.64472e-09 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
RNF31_HUMAN
|
||||||
| NC score | 0.231959 (rank : 8) | θ value | 1.43324e-05 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
RNF31_MOUSE
|
||||||
| NC score | 0.227783 (rank : 9) | θ value | 1.87187e-05 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q924T7 | Gene names | Rnf31 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31. | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.196760 (rank : 10) | θ value | 6.00763e-12 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
YAF2_MOUSE
|
||||||
| NC score | 0.195626 (rank : 11) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LW6, Q3UFD4, Q9DBE4 | Gene names | Yaf2 | |||
|
Domain Architecture |

|
|||||
| Description | YY1-associated factor 2. | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.191855 (rank : 12) | θ value | 1.25267e-09 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
YAF2_HUMAN
|
||||||
| NC score | 0.190989 (rank : 13) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IY57, Q99710 | Gene names | YAF2 | |||
|
Domain Architecture |

|
|||||
| Description | YY1-associated factor 2. | |||||
|
RYBP_HUMAN
|
||||||
| NC score | 0.176625 (rank : 14) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N488, Q9P2W5, Q9UMW4 | Gene names | RYBP, DEDAF, YEAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING1 and YY1-binding protein (Death effector domain-associated factor) (DED-associated factor) (YY1 and E4TF1-associated factor 1) (Apoptin-associating protein 1) (APAP-1). | |||||
|
RYBP_MOUSE
|
||||||
| NC score | 0.173368 (rank : 15) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CCI5, Q9WVK2 | Gene names | Rybp, Dedaf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING1 and YY1-binding protein (Death effector domain-associated factor) (DED-associated factor). | |||||
|
NEIL3_MOUSE
|
||||||
| NC score | 0.150177 (rank : 16) | θ value | 0.00869519 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL3_HUMAN
|
||||||
| NC score | 0.149190 (rank : 17) | θ value | 0.0148317 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
TAB2_HUMAN
|
||||||
| NC score | 0.136584 (rank : 18) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.131888 (rank : 19) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
RBP22_HUMAN
|
||||||
| NC score | 0.118589 (rank : 20) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RGPD8_HUMAN
|
||||||
| NC score | 0.118380 (rank : 21) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
RBP26_HUMAN
|
||||||
| NC score | 0.118302 (rank : 22) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.118010 (rank : 23) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.114005 (rank : 24) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
ZRAB2_HUMAN
|
||||||
| NC score | 0.112546 (rank : 25) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
VCIP1_MOUSE
|
||||||
| NC score | 0.112080 (rank : 26) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CDG3, Q7TMU9, Q8BP90 | Gene names | Vcpip1, Vcip135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
VCIP1_HUMAN
|
||||||
| NC score | 0.111951 (rank : 27) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JH7, Q504T4, Q86T93, Q86W01, Q8N3A9, Q9H5R8 | Gene names | VCPIP1, KIAA1850, VCIP135 | |||
|
Domain Architecture |

|
|||||
| Description | Deubiquitinating protein VCIP135 (EC 3.4.22.-) (Valosin-containing protein p97/p47 complex-interacting protein p135) (Valosin-containing protein p97/p47 complex-interacting protein 1). | |||||
|
RANG_MOUSE
|
||||||
| NC score | 0.111029 (rank : 28) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34022, P34023, Q3TL81, Q9D894, Q9DCA3 | Gene names | Ranbp1, Htf9-a, Htf9a | |||
|
Domain Architecture |

|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RANBP1) (HpaII tiny fragments locus 9a protein). | |||||
|
RANG_HUMAN
|
||||||
| NC score | 0.110245 (rank : 29) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43487 | Gene names | RANBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RanBP1). | |||||
|
NPL4_MOUSE
|
||||||
| NC score | 0.084139 (rank : 30) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P60670 | Gene names | Nploc4, Kiaa1499, Npl4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear protein localization protein 4 homolog (Protein NPL4). | |||||
|
NPL4_HUMAN
|
||||||
| NC score | 0.084066 (rank : 31) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TAT6, Q8N3J1, Q9H8V2, Q9H964, Q9NWR5, Q9P229 | Gene names | NPLOC4, KIAA1499, NPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear protein localization protein 4 homolog (Protein NPL4). | |||||
|
R113B_HUMAN
|
||||||
| NC score | 0.068405 (rank : 32) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
ZFAN6_HUMAN
|
||||||
| NC score | 0.067501 (rank : 33) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6FIF0, O95792, Q9BQF7, Q9GZY3 | Gene names | ZFAND6, AWP1, ZA20D3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 6 (Zinc finger A20 domain-containing protein 3) (Associated with PRK1 protein). | |||||
|
PPIF_MOUSE
|
||||||
| NC score | 0.066958 (rank : 34) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KR7 | Gene names | Ppif | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
PPIA_MOUSE
|
||||||
| NC score | 0.066306 (rank : 35) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17742, Q9CWJ5, Q9R137 | Gene names | Ppia | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein) (SP18). | |||||
|
PPIA_HUMAN
|
||||||
| NC score | 0.066256 (rank : 36) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P62937, P05092, Q6IBU5, Q96IX3, Q9BRU4, Q9BTY9, Q9UC61 | Gene names | PPIA, CYPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase A) (Rotamase A) (Cyclophilin A) (Cyclosporin A-binding protein). | |||||
|
PPIF_HUMAN
|
||||||
| NC score | 0.065896 (rank : 37) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30405, Q5W131 | Gene names | PPIF, CYP3 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase, mitochondrial precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin F). | |||||
|
ZFAN5_HUMAN
|
||||||
| NC score | 0.065882 (rank : 38) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O76080 | Gene names | ZFAND5, ZA20D2, ZNF216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
ZFAN5_MOUSE
|
||||||
| NC score | 0.065233 (rank : 39) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88878 | Gene names | Zfand5, Za20d2, Zfp216, Znf216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
SPN90_MOUSE
|
||||||
| NC score | 0.062974 (rank : 40) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESJ4, Q68G72 | Gene names | Nckipsd, Spin90, Wasbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (N-WASP-interacting protein, 90kDa) (Wiskott-Aldrich syndrome protein-binding protein WISH) (N-WASP-binding protein). | |||||
|
ZFAN6_MOUSE
|
||||||
| NC score | 0.062136 (rank : 41) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCH6, Q9ER79 | Gene names | Zfand6, Awp1, Za20d3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 6 (Zinc finger A20 domain-containing protein 3) (Associated with PRK1 protein). | |||||
|
R113A_HUMAN
|
||||||
| NC score | 0.060321 (rank : 42) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
PPIH_MOUSE
|
||||||
| NC score | 0.059617 (rank : 43) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D868, Q9CQU7 | Gene names | Ppih | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H). | |||||
|
PPIH_HUMAN
|
||||||
| NC score | 0.059106 (rank : 44) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43447 | Gene names | PPIH, CYP20, CYPH | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase H (EC 5.2.1.8) (PPIase H) (Rotamase H) (U-snRNP-associated cyclophilin SnuCyp-20) (USA-CYP) (Small nuclear ribonucleoprotein particle-specific cyclophilin H) (CypH). | |||||
|
PPIB_HUMAN
|
||||||
| NC score | 0.058811 (rank : 45) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23284, Q9BVK5 | Gene names | PPIB, CYPB | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIB_MOUSE
|
||||||
| NC score | 0.058735 (rank : 46) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24369 | Gene names | Ppib | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase B precursor (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin B) (S-cyclophilin) (SCYLP) (CYP-S1). | |||||
|
PPIC_HUMAN
|
||||||
| NC score | 0.058701 (rank : 47) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P45877 | Gene names | PPIC, CYPC | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
PPIC_MOUSE
|
||||||
| NC score | 0.058329 (rank : 48) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30412 | Gene names | Ppic, Cypc | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase C (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin C). | |||||
|
SPN90_HUMAN
|
||||||
| NC score | 0.057117 (rank : 49) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZQ3, Q9UGM8 | Gene names | NCKIPSD, AF3P21, SPIN90 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) (SH3 protein interacting with Nck, 90 kDa) (VacA-interacting protein, 54 kDa) (VIP54) (AF3p21) (Diaphanous protein-interacting protein) (Dia-interacting protein 1) (DIP-1). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.053068 (rank : 50) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.052684 (rank : 51) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPIE_MOUSE
|
||||||
| NC score | 0.052639 (rank : 52) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZH3, Q9D8C0 | Gene names | Ppie, Cyp33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
PPIE_HUMAN
|
||||||
| NC score | 0.052593 (rank : 53) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNP9, O43634, O43635, Q5TGA0, Q9UIZ5 | Gene names | PPIE, CYP33 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase E (EC 5.2.1.8) (PPIase E) (Rotamase E) (Cyclophilin E) (Cyclophilin 33). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.052137 (rank : 54) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.025135 (rank : 55) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.024854 (rank : 56) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
ELOA2_HUMAN
|
||||||
| NC score | 0.023174 (rank : 57) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
SRGP1_HUMAN
|
||||||
| NC score | 0.022083 (rank : 58) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z6B7, Q9H8A3, Q9P2P2 | Gene names | SRGAP1, ARHGAP13, KIAA1304 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 1 (srGAP1) (Rho-GTPase- activating protein 13). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.017676 (rank : 59) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.016685 (rank : 60) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.016282 (rank : 61) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.016256 (rank : 62) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
MUG2_MOUSE
|
||||||
| NC score | 0.014962 (rank : 63) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28666 | Gene names | Mug2, Mug-2 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-2 precursor (MuG2). | |||||
|
MUG1_MOUSE
|
||||||
| NC score | 0.014572 (rank : 64) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28665 | Gene names | Mug1, Mug-1 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-1 precursor (MuG1). | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.013696 (rank : 65) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.009955 (rank : 66) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
GASP1_HUMAN
|
||||||
| NC score | 0.009352 (rank : 67) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.008977 (rank : 68) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
UDA1_HUMAN
|
||||||
| NC score | 0.007267 (rank : 69) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4X1 | Gene names | UGT2A1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 2A1 precursor (EC 2.4.1.17). | |||||
|
CDKL3_MOUSE
|
||||||
| NC score | -0.001513 (rank : 70) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BLF2, Q5M6W2, Q8BKR2, Q8BL49, Q8BVE0, Q8K134 | Gene names | Cdkl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 3 (EC 2.7.11.22). | |||||