Please be patient as the page loads
|
NEIL3_HUMAN
|
||||||
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NEIL3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982026 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
TOP3A_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 3) | NC score | 0.324149 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13472, Q13473 | Gene names | TOP3A, TOP3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
TOP3A_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 4) | NC score | 0.324858 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
NEIL2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 5) | NC score | 0.416445 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969S2, Q7Z3Q7, Q8N842, Q8NG52 | Gene names | NEIL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
NEIL2_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 6) | NC score | 0.377095 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6R2P8 | Gene names | Neil2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
APEX2_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 7) | NC score | 0.319748 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
APEX2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 8) | NC score | 0.307134 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 9) | NC score | 0.096930 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 10) | NC score | 0.116157 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
TTF2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 11) | NC score | 0.105243 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
ZRAN1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.149190 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
RBP2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.089928 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
UB7I3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.098718 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYM8, O95623, Q96BS3, Q9BYM9 | Gene names | RBCK1, C20orf18, RNF54, UBCE7IP3, XAP4 | |||
|
Domain Architecture |
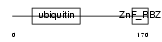
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (Hepatitis B virus X- associated protein 4) (HBV-associated factor 4) (RING finger protein 54). | |||||
|
CASC5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.059019 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
UB7I3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.091203 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |
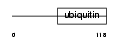
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.040854 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.041046 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.030808 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
ABCA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.016016 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78363, O15112, O60438, O60915 | Gene names | ABCA4, ABCR | |||
|
Domain Architecture |

|
|||||
| Description | Retinal-specific ATP-binding cassette transporter (ATP-binding cassette sub-family A member 4) (RIM ABC transporter) (RIM protein) (RmP) (Stargardt disease protein). | |||||
|
FANCJ_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.035882 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SXJ3, Q8BJQ8, Q8BKI6 | Gene names | Brip1, Bach1, Fancj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
UBAP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.041177 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ09, Q4V759, Q5T7B3, Q6FI75, Q8NC52, Q8NCG6, Q8NCH9 | Gene names | UBAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 1 (UBAP). | |||||
|
ZCHC4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.093358 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H5U6, Q5IW78, Q96AN7 | Gene names | ZCCHC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
ZCHC4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.111218 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BKW4, Q3UTX6, Q9D2I1 | Gene names | Zcchc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.013290 (rank : 46) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
MBD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.023075 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2E2, Q792D6, Q8CCL9, Q9DC19 | Gene names | Mbd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.082063 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.027645 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.007205 (rank : 47) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
JHD1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.014762 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHM5, Q8NCI2, Q96HC7, Q96SL0, Q96T03, Q9NS96, Q9UF75 | Gene names | FBXL10, CXXC2, FBL10, JHDM1B, PCCX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10) (Protein JEMMA) (Jumonji domain-containing EMSY-interactor methyltransferase motif protein) (CXXC-type zinc finger protein 2) (Protein-containing CXXC domain 2). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.014750 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
MDM4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.032783 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15151, Q6GS18, Q8IV83 | Gene names | MDM4, MDMX | |||
|
Domain Architecture |
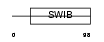
|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
MDM4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.034807 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35618 | Gene names | Mdm4, Mdmx | |||
|
Domain Architecture |
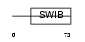
|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
MK08_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.000350 (rank : 50) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 834 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P45983, Q15709, Q15712, Q15713 | Gene names | MAPK8, JNK1, PRKM8 | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase 8 (EC 2.7.11.24) (Stress-activated protein kinase JNK1) (c-Jun N-terminal kinase 1) (JNK-46). | |||||
|
PTPRO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.005711 (rank : 48) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
RYBP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.034313 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N488, Q9P2W5, Q9UMW4 | Gene names | RYBP, DEDAF, YEAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING1 and YY1-binding protein (Death effector domain-associated factor) (DED-associated factor) (YY1 and E4TF1-associated factor 1) (Apoptin-associating protein 1) (APAP-1). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.014767 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.001148 (rank : 49) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
K0423_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.018845 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
K0423_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.019350 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.024639 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
NEIL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.079399 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96FI4, Q6ZRA7, Q86XW7, Q9H6C3 | Gene names | NEIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1) (FPG1). | |||||
|
NEIL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.058258 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
RANG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.050601 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43487 | Gene names | RANBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RanBP1). | |||||
|
RANG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.050985 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34022, P34023, Q3TL81, Q9D894, Q9DCA3 | Gene names | Ranbp1, Htf9-a, Htf9a | |||
|
Domain Architecture |
|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RANBP1) (HpaII tiny fragments locus 9a protein). | |||||
|
RBP22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.052856 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RBP26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.053049 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
RGPD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.052999 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
TOP3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.104820 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95985, Q9BUP5 | Gene names | TOP3B, TOP3B1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
TOP3B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.105161 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z321 | Gene names | Top3b, Top3b1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
NEIL3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL3_MOUSE
|
||||||
| NC score | 0.982026 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K203, Q8CD85, Q8R3P4 | Gene names | Neil3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
NEIL2_HUMAN
|
||||||
| NC score | 0.416445 (rank : 3) | θ value | 1.38499e-08 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969S2, Q7Z3Q7, Q8N842, Q8NG52 | Gene names | NEIL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
NEIL2_MOUSE
|
||||||
| NC score | 0.377095 (rank : 4) | θ value | 1.29631e-06 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6R2P8 | Gene names | Neil2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 2 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 2) (DNA glycosylase/AP lyase Neil2) (DNA-(apurinic or apyrimidinic site) lyase Neil2) (NEH2). | |||||
|
TOP3A_MOUSE
|
||||||
| NC score | 0.324858 (rank : 5) | θ value | 8.11959e-09 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
TOP3A_HUMAN
|
||||||
| NC score | 0.324149 (rank : 6) | θ value | 8.11959e-09 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13472, Q13473 | Gene names | TOP3A, TOP3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
APEX2_MOUSE
|
||||||
| NC score | 0.319748 (rank : 7) | θ value | 6.43352e-06 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68G58, Q8BJP7, Q8BTR7, Q8BUZ2, Q8BYE9, Q8R018, Q8R328, Q9CS12 | Gene names | Apex2, Ape2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2). | |||||
|
APEX2_HUMAN
|
||||||
| NC score | 0.307134 (rank : 8) | θ value | 2.44474e-05 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBZ4, Q9Y5X7 | Gene names | APEX2, APE2, APEXL2, XTH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-(apurinic or apyrimidinic site) lyase 2 (EC 4.2.99.18) (Apurinic- apyrimidinic endonuclease 2) (AP endonuclease 2) (APEX nuclease 2) (APEX nuclease-like 2) (AP endonuclease XTH2). | |||||
|
ZRAN1_HUMAN
|
||||||
| NC score | 0.149190 (rank : 9) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.116157 (rank : 10) | θ value | 0.000786445 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ZCHC4_MOUSE
|
||||||
| NC score | 0.111218 (rank : 11) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BKW4, Q3UTX6, Q9D2I1 | Gene names | Zcchc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
TTF2_HUMAN
|
||||||
| NC score | 0.105243 (rank : 12) | θ value | 0.00298849 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
TOP3B_MOUSE
|
||||||
| NC score | 0.105161 (rank : 13) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z321 | Gene names | Top3b, Top3b1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
TOP3B_HUMAN
|
||||||
| NC score | 0.104820 (rank : 14) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95985, Q9BUP5 | Gene names | TOP3B, TOP3B1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-beta-1 (EC 5.99.1.2) (DNA topoisomerase III beta- 1). | |||||
|
UB7I3_HUMAN
|
||||||
| NC score | 0.098718 (rank : 15) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYM8, O95623, Q96BS3, Q9BYM9 | Gene names | RBCK1, C20orf18, RNF54, UBCE7IP3, XAP4 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (Hepatitis B virus X- associated protein 4) (HBV-associated factor 4) (RING finger protein 54). | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.096930 (rank : 16) | θ value | 0.000158464 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
ZCHC4_HUMAN
|
||||||
| NC score | 0.093358 (rank : 17) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H5U6, Q5IW78, Q96AN7 | Gene names | ZCCHC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 4. | |||||
|
UB7I3_MOUSE
|
||||||
| NC score | 0.091203 (rank : 18) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
RBP2_MOUSE
|
||||||
| NC score | 0.089928 (rank : 19) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERU9, Q61992, Q8C9K9 | Gene names | Ranbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2 (RanBP2). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.082063 (rank : 20) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
NEIL1_HUMAN
|
||||||
| NC score | 0.079399 (rank : 21) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96FI4, Q6ZRA7, Q86XW7, Q9H6C3 | Gene names | NEIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1) (FPG1). | |||||
|
CASC5_MOUSE
|
||||||
| NC score | 0.059019 (rank : 22) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
NEIL1_MOUSE
|
||||||
| NC score | 0.058258 (rank : 23) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4Q6, Q80V58, Q9CYT9 | Gene names | Neil1, Nei1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 1 (EC 3.2.2.-) (EC 4.2.99.18) (Nei-like 1) (DNA glycosylase/AP lyase Neil1) (DNA-(apurinic or apyrimidinic site) lyase Neil1) (NEH1). | |||||
|
RBP26_HUMAN
|
||||||
| NC score | 0.053049 (rank : 24) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
RGPD8_HUMAN
|
||||||
| NC score | 0.052999 (rank : 25) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
RBP22_HUMAN
|
||||||
| NC score | 0.052856 (rank : 26) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q53T03, Q53QN2, Q59GM7 | Gene names | RANBP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 2 (RanBP2L2). | |||||
|
RANG_MOUSE
|
||||||
| NC score | 0.050985 (rank : 27) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34022, P34023, Q3TL81, Q9D894, Q9DCA3 | Gene names | Ranbp1, Htf9-a, Htf9a | |||
|
Domain Architecture |

|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RANBP1) (HpaII tiny fragments locus 9a protein). | |||||
|
RANG_HUMAN
|
||||||
| NC score | 0.050601 (rank : 28) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43487 | Gene names | RANBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-specific GTPase-activating protein (Ran-binding protein 1) (RanBP1). | |||||
|
UBAP1_HUMAN
|
||||||
| NC score | 0.041177 (rank : 29) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ09, Q4V759, Q5T7B3, Q6FI75, Q8NC52, Q8NCG6, Q8NCH9 | Gene names | UBAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 1 (UBAP). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.041046 (rank : 30) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.040854 (rank : 31) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
FANCJ_MOUSE
|
||||||
| NC score | 0.035882 (rank : 32) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SXJ3, Q8BJQ8, Q8BKI6 | Gene names | Brip1, Bach1, Fancj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
MDM4_MOUSE
|
||||||
| NC score | 0.034807 (rank : 33) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35618 | Gene names | Mdm4, Mdmx | |||
|
Domain Architecture |

|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
RYBP_HUMAN
|
||||||
| NC score | 0.034313 (rank : 34) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N488, Q9P2W5, Q9UMW4 | Gene names | RYBP, DEDAF, YEAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING1 and YY1-binding protein (Death effector domain-associated factor) (DED-associated factor) (YY1 and E4TF1-associated factor 1) (Apoptin-associating protein 1) (APAP-1). | |||||
|
MDM4_HUMAN
|
||||||
| NC score | 0.032783 (rank : 35) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15151, Q6GS18, Q8IV83 | Gene names | MDM4, MDMX | |||
|
Domain Architecture |

|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.030808 (rank : 36) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.027645 (rank : 37) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.024639 (rank : 38) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
MBD1_MOUSE
|
||||||
| NC score | 0.023075 (rank : 39) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2E2, Q792D6, Q8CCL9, Q9DC19 | Gene names | Mbd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 1 (Methyl-CpG-binding protein MBD1). | |||||
|
K0423_MOUSE
|
||||||
| NC score | 0.019350 (rank : 40) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.018845 (rank : 41) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
ABCA4_HUMAN
|
||||||
| NC score | 0.016016 (rank : 42) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78363, O15112, O60438, O60915 | Gene names | ABCA4, ABCR | |||
|
Domain Architecture |

|
|||||
| Description | Retinal-specific ATP-binding cassette transporter (ATP-binding cassette sub-family A member 4) (RIM ABC transporter) (RIM protein) (RmP) (Stargardt disease protein). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.014767 (rank : 43) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
JHD1B_HUMAN
|
||||||
| NC score | 0.014762 (rank : 44) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHM5, Q8NCI2, Q96HC7, Q96SL0, Q96T03, Q9NS96, Q9UF75 | Gene names | FBXL10, CXXC2, FBL10, JHDM1B, PCCX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10) (Protein JEMMA) (Jumonji domain-containing EMSY-interactor methyltransferase motif protein) (CXXC-type zinc finger protein 2) (Protein-containing CXXC domain 2). | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.014750 (rank : 45) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.013290 (rank : 46) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.007205 (rank : 47) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PTPRO_HUMAN
|
||||||
| NC score | 0.005711 (rank : 48) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.001148 (rank : 49) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
MK08_HUMAN
|
||||||
| NC score | 0.000350 (rank : 50) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 834 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P45983, Q15709, Q15712, Q15713 | Gene names | MAPK8, JNK1, PRKM8 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 8 (EC 2.7.11.24) (Stress-activated protein kinase JNK1) (c-Jun N-terminal kinase 1) (JNK-46). | |||||