Please be patient as the page loads
|
USP9X_MOUSE
|
||||||
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
USP9X_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998198 (rank : 2) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 171 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.994715 (rank : 3) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 4.52334e-185 (rank : 4) | NC score | 0.932613 (rank : 4) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 1.90147e-66 (rank : 5) | NC score | 0.806638 (rank : 6) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 1.2325e-65 (rank : 6) | NC score | 0.806973 (rank : 5) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | 3.3877e-31 (rank : 7) | NC score | 0.776188 (rank : 8) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 8) | NC score | 0.756514 (rank : 11) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 9) | NC score | 0.772566 (rank : 10) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 10) | NC score | 0.753572 (rank : 12) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 11) | NC score | 0.782991 (rank : 7) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 12) | NC score | 0.772734 (rank : 9) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 13) | NC score | 0.616122 (rank : 37) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 14) | NC score | 0.648673 (rank : 28) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 15) | NC score | 0.693640 (rank : 19) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 16) | NC score | 0.715011 (rank : 15) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UB17L_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 17) | NC score | 0.698266 (rank : 17) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 18) | NC score | 0.716360 (rank : 13) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 19) | NC score | 0.716360 (rank : 14) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 20) | NC score | 0.657958 (rank : 26) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 21) | NC score | 0.671946 (rank : 20) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 22) | NC score | 0.670359 (rank : 22) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 23) | NC score | 0.659759 (rank : 24) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 24) | NC score | 0.659473 (rank : 25) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 25) | NC score | 0.663577 (rank : 23) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 26) | NC score | 0.646166 (rank : 29) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 27) | NC score | 0.631254 (rank : 31) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 28) | NC score | 0.693882 (rank : 18) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP20_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 29) | NC score | 0.652802 (rank : 27) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 30) | NC score | 0.628022 (rank : 34) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 31) | NC score | 0.558561 (rank : 44) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 32) | NC score | 0.629282 (rank : 33) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 33) | NC score | 0.558442 (rank : 45) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 34) | NC score | 0.707125 (rank : 16) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 35) | NC score | 0.631089 (rank : 32) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 36) | NC score | 0.572375 (rank : 41) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 37) | NC score | 0.558716 (rank : 43) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 38) | NC score | 0.627814 (rank : 35) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 39) | NC score | 0.541484 (rank : 50) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 40) | NC score | 0.670948 (rank : 21) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 41) | NC score | 0.645946 (rank : 30) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 42) | NC score | 0.555236 (rank : 46) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 43) | NC score | 0.472004 (rank : 58) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 44) | NC score | 0.471263 (rank : 59) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 45) | NC score | 0.538301 (rank : 52) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 46) | NC score | 0.549709 (rank : 48) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 47) | NC score | 0.610652 (rank : 38) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 48) | NC score | 0.594202 (rank : 40) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 49) | NC score | 0.608805 (rank : 39) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 50) | NC score | 0.621819 (rank : 36) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 51) | NC score | 0.537326 (rank : 53) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 52) | NC score | 0.552981 (rank : 47) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP14_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 53) | NC score | 0.517489 (rank : 56) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP25_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 54) | NC score | 0.449796 (rank : 64) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP25_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 55) | NC score | 0.450933 (rank : 63) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 56) | NC score | 0.541326 (rank : 51) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 57) | NC score | 0.534718 (rank : 54) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 58) | NC score | 0.533549 (rank : 55) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 59) | NC score | 0.463275 (rank : 60) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 60) | NC score | 0.453644 (rank : 62) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 61) | NC score | 0.547390 (rank : 49) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 62) | NC score | 0.485707 (rank : 57) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 63) | NC score | 0.559494 (rank : 42) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 64) | NC score | 0.427352 (rank : 65) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 65) | NC score | 0.396172 (rank : 69) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 66) | NC score | 0.398272 (rank : 67) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 67) | NC score | 0.377098 (rank : 71) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 68) | NC score | 0.381735 (rank : 70) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 69) | NC score | 0.062289 (rank : 78) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 70) | NC score | 0.332182 (rank : 73) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 71) | NC score | 0.397872 (rank : 68) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 72) | NC score | 0.022772 (rank : 99) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 73) | NC score | 0.408163 (rank : 66) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 74) | NC score | 0.048162 (rank : 81) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
CCDC9_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 75) | NC score | 0.055527 (rank : 80) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
IMMT_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 76) | NC score | 0.021925 (rank : 100) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16891, Q14539, Q15092, Q68D41, Q69HW5, Q6IBL0, Q7Z3X1, Q8TAJ5, Q9P0V2 | Gene names | IMMT, HMP, PIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin) (p87/89) (Proliferation-inducing gene 4 protein). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 0.163984 (rank : 77) | NC score | 0.047752 (rank : 82) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
UBP29_MOUSE
|
||||||
| θ value | 0.163984 (rank : 78) | NC score | 0.376220 (rank : 72) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 0.279714 (rank : 79) | NC score | 0.092917 (rank : 75) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
CHRC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 80) | NC score | 0.069656 (rank : 77) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRG0 | Gene names | CHRAC1, CHRAC15 | |||
|
Domain Architecture |
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (CHRAC-15) (HuCHRAC15) (DNA polymerase epsilon subunit p15). | |||||
|
NEIL3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 81) | NC score | 0.041046 (rank : 84) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 0.365318 (rank : 82) | NC score | 0.029172 (rank : 92) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 83) | NC score | 0.016247 (rank : 107) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 84) | NC score | 0.046042 (rank : 83) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
PRP5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 85) | NC score | 0.097307 (rank : 74) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 86) | NC score | 0.014057 (rank : 111) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SYN3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 87) | NC score | 0.027818 (rank : 93) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14994 | Gene names | SYN3 | |||
|
Domain Architecture |
|
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.023385 (rank : 98) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 89) | NC score | 0.016645 (rank : 104) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
ACK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 90) | NC score | -0.001165 (rank : 169) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.010386 (rank : 127) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAF1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.010740 (rank : 125) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0N7 | Gene names | Chaf1b | |||
|
Domain Architecture |
|
|||||
| Description | Chromatin assembly factor 1 subunit B (CAF-1 subunit B) (Chromatin assembly factor I p60 subunit) (CAF-I 60 kDa subunit) (CAF-Ip60). | |||||
|
LIX1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.031058 (rank : 91) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P566, Q8VHY1, Q9CYH4 | Gene names | Lix1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein limb expression 1 homolog. | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.008485 (rank : 137) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.026233 (rank : 94) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.013020 (rank : 118) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.002763 (rank : 158) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
BCL7C_MOUSE
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.031266 (rank : 89) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
BIRC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.031155 (rank : 90) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.009129 (rank : 134) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SGOL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.025687 (rank : 95) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CXH7, Q3U4K4, Q588H1, Q8BKW2 | Gene names | Sgol1, Sgo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1. | |||||
|
CU013_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.020165 (rank : 102) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.019432 (rank : 103) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
AN13B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.007916 (rank : 142) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5F259, Q6IR40 | Gene names | Ankrd13b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13B. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.009395 (rank : 133) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CI126_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.034595 (rank : 85) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N9R8, Q5T163, Q8N1I4 | Gene names | C9orf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf126. | |||||
|
CI126_MOUSE
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.034579 (rank : 86) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C8N2, Q8C409, Q8C8K2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf126 homolog. | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.008126 (rank : 141) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.015790 (rank : 108) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
KIFC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.007133 (rank : 146) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AC6 | Gene names | KIFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC2. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.008865 (rank : 135) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PHF10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.008403 (rank : 139) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
SP6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.000021 (rank : 167) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ESX2, Q6VM22 | Gene names | Sp6, Epfn, Klf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14) (Epiprofin). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.004055 (rank : 152) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
ARY1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.013488 (rank : 113) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50294 | Gene names | Nat1, Aac1 | |||
|
Domain Architecture |

|
|||||
| Description | Arylamine N-acetyltransferase 1 (EC 2.3.1.5) (Arylamide acetylase 1) (N-acetyltransferase type 1) (NAT-1). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.001278 (rank : 163) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BLNK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.010925 (rank : 124) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
CALX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.021680 (rank : 101) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
COBA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.007238 (rank : 145) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.009889 (rank : 128) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
EMIL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.012930 (rank : 119) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.007376 (rank : 144) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.013216 (rank : 115) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.013217 (rank : 114) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
IPP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.024349 (rank : 97) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERT9 | Gene names | Ppp1r1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase inhibitor 1 (IPP-1) (I-1). | |||||
|
KS6C1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | -0.001659 (rank : 172) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLK9, Q3UDY3, Q6PDY4, Q8BMQ5, Q8BX49 | Gene names | Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.009477 (rank : 132) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
MIB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.003669 (rank : 153) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.002376 (rank : 159) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PRR6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.025399 (rank : 96) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z7K6, Q3L8N5, Q8NFH6 | Gene names | PRR6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 6 (Nuclear protein p30). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.000707 (rank : 164) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
TR13C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.031750 (rank : 88) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RJ3 | Gene names | TNFRSF13C, BAFFR, BR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 13C (B cell- activating factor receptor) (BAFF receptor) (BAFF-R) (BLyS receptor 3) (CD268 antigen). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.008298 (rank : 140) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
AMBN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.011072 (rank : 123) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NP70, Q9H2X1, Q9H4L1 | Gene names | AMBN | |||
|
Domain Architecture |
|
|||||
| Description | Ameloblastin precursor. | |||||
|
CF211_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.031916 (rank : 87) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H993, Q96FC6, Q9UFY5 | Gene names | C6orf211 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0364 protein C6orf211. | |||||
|
DONS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.009681 (rank : 129) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYP3, Q9NSR9, Q9NVZ5, Q9NYP1, Q9NYP2 | Gene names | DONSON, C21orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein downstream neighbor of Son (B17). | |||||
|
EMID1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.007583 (rank : 143) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91VF5 | Gene names | Emid1, Emu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
GCM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.013184 (rank : 117) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP62, Q5T0X0, Q99468, Q9P1X3 | Gene names | GCM1, GCMA | |||
|
Domain Architecture |
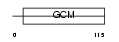
|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (hGCMa). | |||||
|
JADE3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.006554 (rank : 147) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.009602 (rank : 130) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.011259 (rank : 121) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NUFP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.016256 (rank : 106) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5F2E7, Q3TCE2, Q3V195, Q80TF1 | Gene names | Nufip2, Kiaa1321 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 2 (FMRP- interacting protein 2) (82 kDa FMRP-interacting protein) (82-FIP). | |||||
|
PAIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.013214 (rank : 116) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
PDZD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.014037 (rank : 112) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
RTN4R_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.001357 (rank : 161) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZR6 | Gene names | RTN4R, NOGOR | |||
|
Domain Architecture |
|
|||||
| Description | Reticulon-4 receptor precursor (Nogo receptor) (NgR) (Nogo-66 receptor). | |||||
|
RYK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | -0.001206 (rank : 170) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P34925, Q04696 | Gene names | RYK, JTK5A | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase RYK precursor (EC 2.7.10.1). | |||||
|
RYK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | -0.001368 (rank : 171) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01887, Q61890 | Gene names | Ryk, Mrk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase RYK precursor (EC 2.7.10.1) (Kinase VIK) (NYK- R) (Met-related kinase). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.455541 (rank : 61) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.008552 (rank : 136) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
ADA29_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.000053 (rank : 166) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKF5, Q9UHP1, Q9UKF3, Q9UKF4 | Gene names | ADAM29 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 29 precursor (A disintegrin and metalloproteinase domain 29). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.003632 (rank : 154) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAH9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.004540 (rank : 151) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16790 | Gene names | CA9, G250, MN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN) (P54/58N) (Renal cell carcinoma-associated antigen G250) (RCC- associated antigen G250) (pMW1). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.001641 (rank : 160) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
EHD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.005236 (rank : 149) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H223, Q9HAR1, Q9NZN2 | Gene names | EHD4, HCA10, HCA11 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 4 (Hepatocellular carcinoma-associated protein 10/11). | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.011162 (rank : 122) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.014548 (rank : 110) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
H6ST3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.009572 (rank : 131) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.003221 (rank : 156) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HRG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.015325 (rank : 109) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
Domain Architecture |
|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
LRC40_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.000667 (rank : 165) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CRC8, Q78WQ9, Q8BS83 | Gene names | Lrrc40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 40. | |||||
|
LRC8D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | -0.000277 (rank : 168) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGR2, Q3UVA9, Q8CI13 | Gene names | Lrrc8d, Lrrc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.001282 (rank : 162) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
NCOA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.016590 (rank : 105) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13772, Q14239 | Gene names | NCOA4, ARA70, ELE1, RFG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 4 (NCoA-4) (70 kDa androgen receptor coactivator) (70 kDa AR-activator) (Ret-activating protein ELE1). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.010734 (rank : 126) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.012099 (rank : 120) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RPGF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.004921 (rank : 150) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
RPGF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.005459 (rank : 148) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.003283 (rank : 155) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | -0.001879 (rank : 173) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3SY56 | Gene names | SP6, KLF14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.008453 (rank : 138) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
TXLNB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.002821 (rank : 157) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
BRAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.062169 (rank : 79) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
BRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.071361 (rank : 76) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 171 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.998198 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.994715 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.932613 (rank : 4) | θ value | 4.52334e-185 (rank : 4) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.806973 (rank : 5) | θ value | 1.2325e-65 (rank : 6) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.806638 (rank : 6) | θ value | 1.90147e-66 (rank : 5) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.782991 (rank : 7) | θ value | 3.87602e-27 (rank : 11) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.776188 (rank : 8) | θ value | 3.3877e-31 (rank : 7) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.772734 (rank : 9) | θ value | 4.43474e-23 (rank : 12) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.772566 (rank : 10) | θ value | 3.74554e-30 (rank : 9) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.756514 (rank : 11) | θ value | 3.74554e-30 (rank : 8) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.753572 (rank : 12) | θ value | 4.89182e-30 (rank : 10) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.716360 (rank : 13) | θ value | 3.07829e-16 (rank : 18) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.716360 (rank : 14) | θ value | 3.07829e-16 (rank : 19) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.715011 (rank : 15) | θ value | 1.058e-16 (rank : 16) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.707125 (rank : 16) | θ value | 3.89403e-11 (rank : 34) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.698266 (rank : 17) | θ value | 3.07829e-16 (rank : 17) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.693882 (rank : 18) | θ value | 5.43371e-13 (rank : 28) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.693640 (rank : 19) | θ value | 4.74913e-17 (rank : 15) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.671946 (rank : 20) | θ value | 4.44505e-15 (rank : 21) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.670948 (rank : 21) | θ value | 8.11959e-09 (rank : 40) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.670359 (rank : 22) | θ value | 7.58209e-15 (rank : 22) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.663577 (rank : 23) | θ value | 1.86753e-13 (rank : 25) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.659759 (rank : 24) | θ value | 7.58209e-15 (rank : 23) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.659473 (rank : 25) | θ value | 9.90251e-15 (rank : 24) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.657958 (rank : 26) | θ value | 1.99529e-15 (rank : 20) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.652802 (rank : 27) | θ value | 7.09661e-13 (rank : 29) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.648673 (rank : 28) | θ value | 2.7842e-17 (rank : 14) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.646166 (rank : 29) | θ value | 2.43908e-13 (rank : 26) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.645946 (rank : 30) | θ value | 8.11959e-09 (rank : 41) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.631254 (rank : 31) | θ value | 2.43908e-13 (rank : 27) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.631089 (rank : 32) | θ value | 3.29651e-10 (rank : 35) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.629282 (rank : 33) | θ value | 1.33837e-11 (rank : 32) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.628022 (rank : 34) | θ value | 1.2105e-12 (rank : 30) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.627814 (rank : 35) | θ value | 2.79066e-09 (rank : 38) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.621819 (rank : 36) | θ value | 1.53129e-07 (rank : 50) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.616122 (rank : 37) | θ value | 7.32683e-18 (rank : 13) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.610652 (rank : 38) | θ value | 5.26297e-08 (rank : 47) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.608805 (rank : 39) | θ value | 1.53129e-07 (rank : 49) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.594202 (rank : 40) | θ value | 1.53129e-07 (rank : 48) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.572375 (rank : 41) | θ value | 3.29651e-10 (rank : 36) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.559494 (rank : 42) | θ value | 0.000602161 (rank : 63) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.558716 (rank : 43) | θ value | 1.63604e-09 (rank : 37) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.558561 (rank : 44) | θ value | 1.33837e-11 (rank : 31) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.558442 (rank : 45) | θ value | 1.74796e-11 (rank : 33) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.555236 (rank : 46) | θ value | 1.06045e-08 (rank : 42) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.552981 (rank : 47) | θ value | 5.81887e-07 (rank : 52) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.549709 (rank : 48) | θ value | 1.80886e-08 (rank : 46) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.547390 (rank : 49) | θ value | 0.00035302 (rank : 61) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.541484 (rank : 50) | θ value | 4.76016e-09 (rank : 39) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.541326 (rank : 51) | θ value | 1.69304e-06 (rank : 56) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.538301 (rank : 52) | θ value | 1.80886e-08 (rank : 45) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.537326 (rank : 53) | θ value | 4.45536e-07 (rank : 51) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.534718 (rank : 54) | θ value | 1.69304e-06 (rank : 57) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.533549 (rank : 55) | θ value | 1.69304e-06 (rank : 58) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP14_MOUSE
|
||||||
| NC score | 0.517489 (rank : 56) | θ value | 9.92553e-07 (rank : 53) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.485707 (rank : 57) | θ value | 0.00035302 (rank : 62) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.472004 (rank : 58) | θ value | 1.06045e-08 (rank : 43) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_HUMAN
|
||||||
| NC score | 0.471263 (rank : 59) | θ value | 1.38499e-08 (rank : 44) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.463275 (rank : 60) | θ value | 1.43324e-05 (rank : 59) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.455541 (rank : 61) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.453644 (rank : 62) | θ value | 2.44474e-05 (rank : 60) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
UBP25_MOUSE
|
||||||
| NC score | 0.450933 (rank : 63) | θ value | 1.29631e-06 (rank : 55) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP25_HUMAN
|
||||||
| NC score | 0.449796 (rank : 64) | θ value | 9.92553e-07 (rank : 54) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.427352 (rank : 65) | θ value | 0.00175202 (rank : 64) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.408163 (rank : 66) | θ value | 0.0563607 (rank : 73) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.398272 (rank : 67) | θ value | 0.00228821 (rank : 66) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.397872 (rank : 68) | θ value | 0.0193708 (rank : 71) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.396172 (rank : 69) | θ value | 0.00228821 (rank : 65) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.381735 (rank : 70) | θ value | 0.00665767 (rank : 68) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.377098 (rank : 71) | θ value | 0.00509761 (rank : 67) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP29_MOUSE
|
||||||
| NC score | 0.376220 (rank : 72) | θ value | 0.163984 (rank : 78) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.332182 (rank : 73) | θ value | 0.0193708 (rank : 70) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.097307 (rank : 74) | θ value | 0.47712 (rank : 85) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.092917 (rank : 75) | θ value | 0.279714 (rank : 79) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
BRAP_MOUSE
|
||||||
| NC score | 0.071361 (rank : 76) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
CHRC1_HUMAN
|
||||||
| NC score | 0.069656 (rank : 77) | θ value | 0.365318 (rank : 80) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRG0 | Gene names | CHRAC1, CHRAC15 | |||
|
Domain Architecture |

|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (CHRAC-15) (HuCHRAC15) (DNA polymerase epsilon subunit p15). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.062289 (rank : 78) | θ value | 0.0113563 (rank : 69) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
BRAP_HUMAN
|
||||||
| NC score | 0.062169 (rank : 79) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
CCDC9_HUMAN
|
||||||
| NC score | 0.055527 (rank : 80) | θ value | 0.0961366 (rank : 75) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.048162 (rank : 81) | θ value | 0.0736092 (rank : 74) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.047752 (rank : 82) | θ value | 0.163984 (rank : 77) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.046042 (rank : 83) | θ value | 0.47712 (rank : 84) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
NEIL3_HUMAN
|
||||||
| NC score | 0.041046 (rank : 84) | θ value | 0.365318 (rank : 81) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
CI126_HUMAN
|
||||||
| NC score | 0.034595 (rank : 85) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N9R8, Q5T163, Q8N1I4 | Gene names | C9orf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf126. | |||||
|
CI126_MOUSE
|
||||||
| NC score | 0.034579 (rank : 86) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C8N2, Q8C409, Q8C8K2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf126 homolog. | |||||
|
CF211_HUMAN
|
||||||
| NC score | 0.031916 (rank : 87) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H993, Q96FC6, Q9UFY5 | Gene names | C6orf211 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0364 protein C6orf211. | |||||
|
TR13C_HUMAN
|
||||||
| NC score | 0.031750 (rank : 88) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RJ3 | Gene names | TNFRSF13C, BAFFR, BR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 13C (B cell- activating factor receptor) (BAFF receptor) (BAFF-R) (BLyS receptor 3) (CD268 antigen). | |||||
|
BCL7C_MOUSE
|
||||||
| NC score | 0.031266 (rank : 89) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
BIRC6_HUMAN
|
||||||
| NC score | 0.031155 (rank : 90) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NR09, Q9ULD1 | Gene names | BIRC6, KIAA1289 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 6 (Ubiquitin-conjugating BIR-domain enzyme apollon). | |||||
|
LIX1_MOUSE
|
||||||
| NC score | 0.031058 (rank : 91) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P566, Q8VHY1, Q9CYH4 | Gene names | Lix1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein limb expression 1 homolog. | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.029172 (rank : 92) | θ value | 0.365318 (rank : 82) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
SYN3_HUMAN
|
||||||
| NC score | 0.027818 (rank : 93) | θ value | 0.813845 (rank : 87) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14994 | Gene names | SYN3 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.026233 (rank : 94) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
SGOL1_MOUSE
|
||||||
| NC score | 0.025687 (rank : 95) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CXH7, Q3U4K4, Q588H1, Q8BKW2 | Gene names | Sgol1, Sgo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1. | |||||
|
PRR6_HUMAN
|
||||||
| NC score | 0.025399 (rank : 96) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z7K6, Q3L8N5, Q8NFH6 | Gene names | PRR6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 6 (Nuclear protein p30). | |||||
|
IPP1_MOUSE
|
||||||
| NC score | 0.024349 (rank : 97) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERT9 | Gene names | Ppp1r1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase inhibitor 1 (IPP-1) (I-1). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.023385 (rank : 98) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.022772 (rank : 99) | θ value | 0.0252991 (rank : 72) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
IMMT_HUMAN
|
||||||
| NC score | 0.021925 (rank : 100) | θ value | 0.0961366 (rank : 76) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16891, Q14539, Q15092, Q68D41, Q69HW5, Q6IBL0, Q7Z3X1, Q8TAJ5, Q9P0V2 | Gene names | IMMT, HMP, PIG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin) (p87/89) (Proliferation-inducing gene 4 protein). | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.021680 (rank : 101) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CU013_HUMAN
|
||||||
| NC score | 0.020165 (rank : 102) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.019432 (rank : 103) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.016645 (rank : 104) | θ value | 1.06291 (rank : 89) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
NCOA4_HUMAN
|
||||||
| NC score | 0.016590 (rank : 105) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13772, Q14239 | Gene names | NCOA4, ARA70, ELE1, RFG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 4 (NCoA-4) (70 kDa androgen receptor coactivator) (70 kDa AR-activator) (Ret-activating protein ELE1). | |||||
|
NUFP2_MOUSE
|
||||||
| NC score | 0.016256 (rank : 106) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5F2E7, Q3TCE2, Q3V195, Q80TF1 | Gene names | Nufip2, Kiaa1321 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 2 (FMRP- interacting protein 2) (82 kDa FMRP-interacting protein) (82-FIP). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.016247 (rank : 107) | θ value | 0.365318 (rank : 83) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.015790 (rank : 108) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
HRG_HUMAN
|
||||||
| NC score | 0.015325 (rank : 109) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04196 | Gene names | HRG | |||
|
Domain Architecture |

|
|||||
| Description | Histidine-rich glycoprotein precursor (Histidine-proline-rich glycoprotein) (HPRG). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.014548 (rank : 110) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.014057 (rank : 111) | θ value | 0.47712 (rank : 86) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
PDZD8_HUMAN
|
||||||
| NC score | 0.014037 (rank : 112) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEN9, Q86WE0, Q86WE5, Q9UFF1 | Gene names | PDZD8, PDZK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 8 (Sarcoma antigen NY-SAR-84/NY-SAR- 104). | |||||
|
ARY1_MOUSE
|
||||||
| NC score | 0.013488 (rank : 113) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50294 | Gene names | Nat1, Aac1 | |||
|
Domain Architecture |

|
|||||
| Description | Arylamine N-acetyltransferase 1 (EC 2.3.1.5) (Arylamide acetylase 1) (N-acetyltransferase type 1) (NAT-1). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.013217 (rank : 114) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.013216 (rank : 115) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
PAIP1_HUMAN
|
||||||
| NC score | 0.013214 (rank : 116) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
GCM1_HUMAN
|
||||||
| NC score | 0.013184 (rank : 117) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP62, Q5T0X0, Q99468, Q9P1X3 | Gene names | GCM1, GCMA | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (hGCMa). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.013020 (rank : 118) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
EMIL2_MOUSE
|
||||||
| NC score | 0.012930 (rank : 119) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.012099 (rank : 120) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.011259 (rank : 121) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.011162 (rank : 122) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
AMBN_HUMAN
|
||||||
| NC score | 0.011072 (rank : 123) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NP70, Q9H2X1, Q9H4L1 | Gene names | AMBN | |||
|
Domain Architecture |

|
|||||
| Description | Ameloblastin precursor. | |||||
|
BLNK_HUMAN
|
||||||
| NC score | 0.010925 (rank : 124) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
CAF1B_MOUSE
|
||||||
| NC score | 0.010740 (rank : 125) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0N7 | Gene names | Chaf1b | |||
|
Domain Architecture |

|
|||||
| Description | Chromatin assembly factor 1 subunit B (CAF-1 subunit B) (Chromatin assembly factor I p60 subunit) (CAF-I 60 kDa subunit) (CAF-Ip60). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.010734 (rank : 126) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.010386 (rank : 127) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.009889 (rank : 128) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DONS_HUMAN
|
||||||
| NC score | 0.009681 (rank : 129) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYP3, Q9NSR9, Q9NVZ5, Q9NYP1, Q9NYP2 | Gene names | DONSON, C21orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein downstream neighbor of Son (B17). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.009602 (rank : 130) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
H6ST3_MOUSE
|
||||||
| NC score | 0.009572 (rank : 131) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.009477 (rank : 132) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.009395 (rank : 133) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.009129 (rank : 134) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.008865 (rank : 135) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.008552 (rank : 136) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.008485 (rank : 137) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.008453 (rank : 138) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.008403 (rank : 139) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.008298 (rank : 140) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.008126 (rank : 141) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
AN13B_MOUSE
|
||||||
| NC score | 0.007916 (rank : 142) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5F259, Q6IR40 | Gene names | Ankrd13b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13B. | |||||
|
EMID1_MOUSE
|
||||||
| NC score | 0.007583 (rank : 143) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91VF5 | Gene names | Emid1, Emu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMI domain-containing protein 1 precursor (Protein Emu1) (Emilin and multimerin domain-containing protein 1). | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.007376 (rank : 144) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.007238 (rank : 145) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
KIFC2_HUMAN
|
||||||
| NC score | 0.007133 (rank : 146) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AC6 | Gene names | KIFC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC2. | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.006554 (rank : 147) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
RPGF4_MOUSE
|
||||||
| NC score | 0.005459 (rank : 148) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
EHD4_HUMAN
|
||||||
| NC score | 0.005236 (rank : 149) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H223, Q9HAR1, Q9NZN2 | Gene names | EHD4, HCA10, HCA11 | |||
|
Domain Architecture |

|
|||||
| Description | EH-domain-containing protein 4 (Hepatocellular carcinoma-associated protein 10/11). | |||||
|
RPGF4_HUMAN
|
||||||
| NC score | 0.004921 (rank : 150) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
CAH9_HUMAN
|
||||||
| NC score | 0.004540 (rank : 151) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16790 | Gene names | CA9, G250, MN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN) (P54/58N) (Renal cell carcinoma-associated antigen G250) (RCC- associated antigen G250) (pMW1). | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.004055 (rank : 152) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.003669 (rank : 153) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.003632 (rank : 154) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.003283 (rank : 155) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.003221 (rank : 156) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
TXLNB_MOUSE
|
||||||
| NC score | 0.002821 (rank : 157) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.002763 (rank : 158) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.002376 (rank : 159) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.001641 (rank : 160) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
RTN4R_HUMAN
|
||||||
| NC score | 0.001357 (rank : 161) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZR6 | Gene names | RTN4R, NOGOR | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 receptor precursor (Nogo receptor) (NgR) (Nogo-66 receptor). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.001282 (rank : 162) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.001278 (rank : 163) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.000707 (rank : 164) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
LRC40_MOUSE
|
||||||
| NC score | 0.000667 (rank : 165) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CRC8, Q78WQ9, Q8BS83 | Gene names | Lrrc40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 40. | |||||
|
ADA29_HUMAN
|
||||||
| NC score | 0.000053 (rank : 166) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKF5, Q9UHP1, Q9UKF3, Q9UKF4 | Gene names | ADAM29 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 29 precursor (A disintegrin and metalloproteinase domain 29). | |||||
|
SP6_MOUSE
|
||||||
| NC score | 0.000021 (rank : 167) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ESX2, Q6VM22 | Gene names | Sp6, Epfn, Klf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14) (Epiprofin). | |||||
|
LRC8D_MOUSE
|
||||||
| NC score | -0.000277 (rank : 168) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGR2, Q3UVA9, Q8CI13 | Gene names | Lrrc8d, Lrrc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8D. | |||||
|
ACK1_HUMAN
|
||||||
| NC score | -0.001165 (rank : 169) | θ value | 1.38821 (rank : 90) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
RYK_HUMAN
|
||||||
| NC score | -0.001206 (rank : 170) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P34925, Q04696 | Gene names | RYK, JTK5A | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase RYK precursor (EC 2.7.10.1). | |||||
|
RYK_MOUSE
|
||||||
| NC score | -0.001368 (rank : 171) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01887, Q61890 | Gene names | Ryk, Mrk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase RYK precursor (EC 2.7.10.1) (Kinase VIK) (NYK- R) (Met-related kinase). | |||||
|
KS6C1_MOUSE
|
||||||
| NC score | -0.001659 (rank : 172) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLK9, Q3UDY3, Q6PDY4, Q8BMQ5, Q8BX49 | Gene names | Rps6kc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal protein S6 kinase delta-1 (EC 2.7.11.1) (52 kDa ribosomal protein S6 kinase). | |||||
|
SP6_HUMAN
|
||||||
| NC score | -0.001879 (rank : 173) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 171 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3SY56 | Gene names | SP6, KLF14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp6 (Krueppel-like factor 14). | |||||