Please be patient as the page loads
|
UBP25_HUMAN
|
||||||
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBP25_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP25_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994649 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP28_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.974591 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.966771 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 5) | NC score | 0.585316 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 6) | NC score | 0.581865 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 7) | NC score | 0.492418 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 8) | NC score | 0.536544 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 9) | NC score | 0.491152 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 10) | NC score | 0.461587 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 11) | NC score | 0.420795 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 12) | NC score | 0.490010 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 13) | NC score | 0.486854 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 14) | NC score | 0.486854 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 15) | NC score | 0.490992 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 16) | NC score | 0.491565 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 17) | NC score | 0.457512 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 18) | NC score | 0.469797 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 19) | NC score | 0.469961 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 20) | NC score | 0.449796 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 21) | NC score | 0.484858 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 22) | NC score | 0.468892 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 23) | NC score | 0.477483 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 24) | NC score | 0.483497 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 25) | NC score | 0.411085 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 26) | NC score | 0.408513 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 27) | NC score | 0.387974 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 28) | NC score | 0.390700 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 29) | NC score | 0.404974 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 30) | NC score | 0.394454 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 31) | NC score | 0.483725 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 32) | NC score | 0.395503 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 33) | NC score | 0.391408 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 34) | NC score | 0.391089 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP14_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 35) | NC score | 0.384827 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 36) | NC score | 0.385822 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 37) | NC score | 0.379581 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP29_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 38) | NC score | 0.340034 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 39) | NC score | 0.306401 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 40) | NC score | 0.344613 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 41) | NC score | 0.308053 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 42) | NC score | 0.380341 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 43) | NC score | 0.403748 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 44) | NC score | 0.020607 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 45) | NC score | 0.404167 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 46) | NC score | 0.324344 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 47) | NC score | 0.392736 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 48) | NC score | 0.357158 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 49) | NC score | 0.019459 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 50) | NC score | 0.318119 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 51) | NC score | 0.376659 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 52) | NC score | 0.265719 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 53) | NC score | 0.297859 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 54) | NC score | 0.324816 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 55) | NC score | 0.327910 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 56) | NC score | 0.367041 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP20_HUMAN
|
||||||
| θ value | 0.125558 (rank : 57) | NC score | 0.378515 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 0.125558 (rank : 58) | NC score | 0.276779 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UB17L_HUMAN
|
||||||
| θ value | 0.163984 (rank : 59) | NC score | 0.373663 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 0.163984 (rank : 60) | NC score | 0.312415 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 0.163984 (rank : 61) | NC score | 0.312562 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 62) | NC score | 0.350989 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.360211 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 0.21417 (rank : 64) | NC score | 0.358713 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.293670 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 66) | NC score | 0.010484 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.271802 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 68) | NC score | 0.275249 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 69) | NC score | 0.030374 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CAP2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 70) | NC score | 0.046267 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |
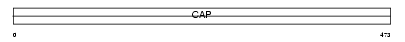
|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.035279 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.035892 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.307989 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
WEE1B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 74) | NC score | 0.006169 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.035265 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.035436 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.023234 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 0.62314 (rank : 78) | NC score | 0.310829 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.009763 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.224873 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.231108 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
DZIP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.025913 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
OSBP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.013334 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.024826 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CAP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.041206 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYT6, Q80YU7, Q9D6L0 | Gene names | Cap2 | |||
|
Domain Architecture |
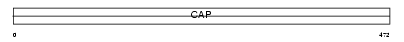
|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | 0.023876 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
RRMJ3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | 0.032980 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.025410 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.015649 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
K1C9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.009696 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.016627 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.354691 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
AT2B3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.013402 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.012746 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.016942 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.016741 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
SNCAP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.011405 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.370801 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.016381 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
SAS6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.019383 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.023393 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.018838 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GUC2C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.003854 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
LYST_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.008306 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
ATX3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.024671 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54252, O15284, O15285, O15286, Q8N189, Q96TC3, Q96TC4, Q9H3N0 | Gene names | ATXN3, ATX3, MJD, MJD1, SCA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-3 (EC 3.4.22.-) (Machado-Joseph disease protein 1) (Spinocerebellar ataxia type 3 protein). | |||||
|
HXB4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.004805 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17483, Q9NTA0 | Gene names | HOXB4, HOX2F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2F) (Hox-2.6). | |||||
|
HXB4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.004826 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10284, Q3UZB4, Q4VBG0 | Gene names | Hoxb4, Hox-2.6, Hoxb-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2.6). | |||||
|
NOL6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.033845 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5K4, Q6PAN8, Q8C6V4, Q8R5K3, Q8VCG0, Q8WTY7 | Gene names | Nol6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
TNIP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.020795 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
ATX3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.026928 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CVD2 | Gene names | Atxn3, Mjd | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-3 (EC 3.4.22.-) (Machado-Joseph disease protein 1 homolog). | |||||
|
ITIH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.006666 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.016172 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.016526 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
PIWL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.009206 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z3Z3 | Gene names | PIWIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 3. | |||||
|
PTC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.007353 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35595, Q546T0 | Gene names | Ptch2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.292334 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
ZN694_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | -0.003758 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.002777 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
PLCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.009466 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PRDM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | -0.002274 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
RFWD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.003337 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
RFWD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.003404 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
RHG25_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.009583 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.018227 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
BRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.050387 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.316467 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP25_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP25_MOUSE
|
||||||
| NC score | 0.994649 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP28_HUMAN
|
||||||
| NC score | 0.974591 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.966771 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.585316 (rank : 5) | θ value | 2.97466e-19 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.581865 (rank : 6) | θ value | 1.92812e-18 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.536544 (rank : 7) | θ value | 3.29651e-10 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.492418 (rank : 8) | θ value | 3.29651e-10 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.491565 (rank : 9) | θ value | 1.99992e-07 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.491152 (rank : 10) | θ value | 4.30538e-10 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.490992 (rank : 11) | θ value | 1.53129e-07 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.490010 (rank : 12) | θ value | 6.87365e-08 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.486854 (rank : 13) | θ value | 6.87365e-08 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.486854 (rank : 14) | θ value | 6.87365e-08 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.484858 (rank : 15) | θ value | 1.69304e-06 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.483725 (rank : 16) | θ value | 0.00134147 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.483497 (rank : 17) | θ value | 4.1701e-05 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.477483 (rank : 18) | θ value | 1.87187e-05 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.469961 (rank : 19) | θ value | 7.59969e-07 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.469797 (rank : 20) | θ value | 5.81887e-07 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.468892 (rank : 21) | θ value | 3.77169e-06 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.461587 (rank : 22) | θ value | 1.06045e-08 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.457512 (rank : 23) | θ value | 1.99992e-07 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.449796 (rank : 24) | θ value | 9.92553e-07 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.420795 (rank : 25) | θ value | 1.38499e-08 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.411085 (rank : 26) | θ value | 9.29e-05 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.408513 (rank : 27) | θ value | 0.000270298 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.404974 (rank : 28) | θ value | 0.000786445 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.404167 (rank : 29) | θ value | 0.0193708 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.403748 (rank : 30) | θ value | 0.0148317 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.395503 (rank : 31) | θ value | 0.00298849 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.394454 (rank : 32) | θ value | 0.000786445 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.392736 (rank : 33) | θ value | 0.0252991 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.391408 (rank : 34) | θ value | 0.00298849 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.391089 (rank : 35) | θ value | 0.00298849 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.390700 (rank : 36) | θ value | 0.000786445 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.387974 (rank : 37) | θ value | 0.000602161 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.385822 (rank : 38) | θ value | 0.00390308 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP14_MOUSE
|
||||||
| NC score | 0.384827 (rank : 39) | θ value | 0.00390308 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.380341 (rank : 40) | θ value | 0.0148317 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.379581 (rank : 41) | θ value | 0.00390308 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.378515 (rank : 42) | θ value | 0.125558 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.376659 (rank : 43) | θ value | 0.0563607 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.373663 (rank : 44) | θ value | 0.163984 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.370801 (rank : 45) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.367041 (rank : 46) | θ value | 0.0961366 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.360211 (rank : 47) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.358713 (rank : 48) | θ value | 0.21417 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.357158 (rank : 49) | θ value | 0.0330416 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.354691 (rank : 50) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.350989 (rank : 51) | θ value | 0.21417 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.344613 (rank : 52) | θ value | 0.00665767 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP29_MOUSE
|
||||||
| NC score | 0.340034 (rank : 53) | θ value | 0.00509761 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.327910 (rank : 54) | θ value | 0.0961366 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.324816 (rank : 55) | θ value | 0.0736092 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.324344 (rank : 56) | θ value | 0.0252991 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.318119 (rank : 57) | θ value | 0.0563607 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.316467 (rank : 58) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.312562 (rank : 59) | θ value | 0.163984 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.312415 (rank : 60) | θ value | 0.163984 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.310829 (rank : 61) | θ value | 0.62314 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.308053 (rank : 62) | θ value | 0.00869519 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.307989 (rank : 63) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.306401 (rank : 64) | θ value | 0.00665767 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.297859 (rank : 65) | θ value | 0.0736092 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.293670 (rank : 66) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.292334 (rank : 67) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.276779 (rank : 68) | θ value | 0.125558 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.275249 (rank : 69) | θ value | 0.365318 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.271802 (rank : 70) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.265719 (rank : 71) | θ value | 0.0563607 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.231108 (rank : 72) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.224873 (rank : 73) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
BRAP_MOUSE
|
||||||
| NC score | 0.050387 (rank : 74) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
CAP2_HUMAN
|
||||||
| NC score | 0.046267 (rank : 75) | θ value | 0.47712 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CAP2_MOUSE
|
||||||
| NC score | 0.041206 (rank : 76) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYT6, Q80YU7, Q9D6L0 | Gene names | Cap2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.035892 (rank : 77) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.035436 (rank : 78) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.035279 (rank : 79) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.035265 (rank : 80) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
NOL6_MOUSE
|
||||||
| NC score | 0.033845 (rank : 81) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5K4, Q6PAN8, Q8C6V4, Q8R5K3, Q8VCG0, Q8WTY7 | Gene names | Nol6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
RRMJ3_MOUSE
|
||||||
| NC score | 0.032980 (rank : 82) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.030374 (rank : 83) | θ value | 0.47712 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
ATX3_MOUSE
|
||||||
| NC score | 0.026928 (rank : 84) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CVD2 | Gene names | Atxn3, Mjd | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-3 (EC 3.4.22.-) (Machado-Joseph disease protein 1 homolog). | |||||
|
DZIP1_MOUSE
|
||||||
| NC score | 0.025913 (rank : 85) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.025410 (rank : 86) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.024826 (rank : 87) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
ATX3_HUMAN
|
||||||
| NC score | 0.024671 (rank : 88) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54252, O15284, O15285, O15286, Q8N189, Q96TC3, Q96TC4, Q9H3N0 | Gene names | ATXN3, ATX3, MJD, MJD1, SCA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-3 (EC 3.4.22.-) (Machado-Joseph disease protein 1) (Spinocerebellar ataxia type 3 protein). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.023876 (rank : 89) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.023393 (rank : 90) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.023234 (rank : 91) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
TNIP3_HUMAN
|
||||||
| NC score | 0.020795 (rank : 92) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.020607 (rank : 93) | θ value | 0.0193708 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.019459 (rank : 94) | θ value | 0.0563607 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SAS6_HUMAN
|
||||||
| NC score | 0.019383 (rank : 95) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.018838 (rank : 96) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.018227 (rank : 97) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.016942 (rank : 98) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.016741 (rank : 99) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.016627 (rank : 100) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.016526 (rank : 101) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.016381 (rank : 102) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.016172 (rank : 103) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.015649 (rank : 104) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
AT2B3_HUMAN
|
||||||
| NC score | 0.013402 (rank : 105) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
OSBP2_MOUSE
|
||||||
| NC score | 0.013334 (rank : 106) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5QNQ6, Q8CF21 | Gene names | Osbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxysterol-binding protein 2. | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.012746 (rank : 107) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
SNCAP_HUMAN
|
||||||
| NC score | 0.011405 (rank : 108) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.010484 (rank : 109) | θ value | 0.365318 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.009763 (rank : 110) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
K1C9_HUMAN
|
||||||
| NC score | 0.009696 (rank : 111) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35527, O00109, Q14665 | Gene names | KRT9 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 9 (Cytokeratin-9) (CK-9) (Keratin-9) (K9). | |||||
|
RHG25_HUMAN
|
||||||
| NC score | 0.009583 (rank : 112) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 550 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P42331, Q8IXQ2 | Gene names | ARHGAP25, KIAA0053 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 25. | |||||
|
PLCB1_HUMAN
|
||||||
| NC score | 0.009466 (rank : 113) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PIWL3_HUMAN
|
||||||
| NC score | 0.009206 (rank : 114) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z3Z3 | Gene names | PIWIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 3. | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.008306 (rank : 115) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
PTC2_MOUSE
|
||||||
| NC score | 0.007353 (rank : 116) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35595, Q546T0 | Gene names | Ptch2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 2 (PTC2). | |||||
|
ITIH2_MOUSE
|
||||||
| NC score | 0.006666 (rank : 117) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61703 | Gene names | Itih2 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H2 precursor (ITI heavy chain H2) (Inter-alpha-inhibitor heavy chain 2). | |||||
|
WEE1B_HUMAN
|
||||||
| NC score | 0.006169 (rank : 118) | θ value | 0.47712 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
HXB4_MOUSE
|
||||||
| NC score | 0.004826 (rank : 119) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10284, Q3UZB4, Q4VBG0 | Gene names | Hoxb4, Hox-2.6, Hoxb-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2.6). | |||||
|
HXB4_HUMAN
|
||||||
| NC score | 0.004805 (rank : 120) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17483, Q9NTA0 | Gene names | HOXB4, HOX2F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B4 (Hox-2F) (Hox-2.6). | |||||
|
GUC2C_HUMAN
|
||||||
| NC score | 0.003854 (rank : 121) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25092 | Gene names | GUCY2C, GUC2C, STAR | |||
|
Domain Architecture |

|
|||||
| Description | Heat-stable enterotoxin receptor precursor (GC-C) (Intestinal guanylate cyclase) (EC 4.6.1.2) (STA receptor) (hSTAR). | |||||
|
RFWD2_MOUSE
|
||||||
| NC score | 0.003404 (rank : 122) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
RFWD2_HUMAN
|
||||||
| NC score | 0.003337 (rank : 123) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.002777 (rank : 124) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
PRDM1_MOUSE
|
||||||
| NC score | -0.002274 (rank : 125) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
ZN694_HUMAN
|
||||||
| NC score | -0.003758 (rank : 126) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63HK3, Q6ZN77 | Gene names | ZNF694 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 694. | |||||