Please be patient as the page loads
|
UBP33_HUMAN
|
||||||
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBP20_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.986072 (rank : 2) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 157 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 3) | NC score | 0.897144 (rank : 4) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 8.91499e-32 (rank : 4) | NC score | 0.891122 (rank : 5) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 8.91499e-32 (rank : 5) | NC score | 0.868023 (rank : 12) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 6) | NC score | 0.826973 (rank : 20) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 7) | NC score | 0.898526 (rank : 3) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 8) | NC score | 0.889741 (rank : 6) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 9) | NC score | 0.871606 (rank : 10) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 10) | NC score | 0.870008 (rank : 11) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 1.47289e-26 (rank : 11) | NC score | 0.873544 (rank : 8) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 12) | NC score | 0.816883 (rank : 31) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 13) | NC score | 0.821838 (rank : 25) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 14) | NC score | 0.826362 (rank : 21) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 15) | NC score | 0.818794 (rank : 28) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | 2.77775e-25 (rank : 16) | NC score | 0.872146 (rank : 9) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 8.08199e-25 (rank : 17) | NC score | 0.882784 (rank : 7) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 18) | NC score | 0.822965 (rank : 24) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 19) | NC score | 0.821331 (rank : 27) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 20) | NC score | 0.821443 (rank : 26) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 21) | NC score | 0.866865 (rank : 13) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 22) | NC score | 0.824807 (rank : 22) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 23) | NC score | 0.797393 (rank : 35) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 24) | NC score | 0.842318 (rank : 15) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 25) | NC score | 0.793824 (rank : 37) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 26) | NC score | 0.831949 (rank : 18) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 27) | NC score | 0.829925 (rank : 19) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 28) | NC score | 0.758026 (rank : 39) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 29) | NC score | 0.785473 (rank : 38) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 30) | NC score | 0.712058 (rank : 41) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 31) | NC score | 0.672412 (rank : 52) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 32) | NC score | 0.838691 (rank : 16) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 33) | NC score | 0.813799 (rank : 32) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 34) | NC score | 0.824002 (rank : 23) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 35) | NC score | 0.853840 (rank : 14) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 36) | NC score | 0.832663 (rank : 17) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 37) | NC score | 0.803398 (rank : 34) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 38) | NC score | 0.794002 (rank : 36) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 39) | NC score | 0.680947 (rank : 49) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 40) | NC score | 0.687834 (rank : 46) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 41) | NC score | 0.818603 (rank : 29) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 42) | NC score | 0.818603 (rank : 30) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 43) | NC score | 0.646717 (rank : 57) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 44) | NC score | 0.646166 (rank : 58) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 45) | NC score | 0.659234 (rank : 56) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UB17L_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 46) | NC score | 0.807246 (rank : 33) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 47) | NC score | 0.659749 (rank : 55) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 48) | NC score | 0.644711 (rank : 59) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 49) | NC score | 0.677595 (rank : 50) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 50) | NC score | 0.673066 (rank : 51) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 51) | NC score | 0.702393 (rank : 44) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 52) | NC score | 0.711747 (rank : 43) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 53) | NC score | 0.711896 (rank : 42) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 54) | NC score | 0.714148 (rank : 40) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 55) | NC score | 0.683049 (rank : 48) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 56) | NC score | 0.669310 (rank : 53) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 57) | NC score | 0.664618 (rank : 54) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 58) | NC score | 0.686686 (rank : 47) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 59) | NC score | 0.519937 (rank : 64) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
BRAP_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 60) | NC score | 0.188087 (rank : 75) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
BRAP_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 61) | NC score | 0.212450 (rank : 74) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 62) | NC score | 0.621194 (rank : 60) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 63) | NC score | 0.427765 (rank : 70) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP14_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 64) | NC score | 0.607134 (rank : 62) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 65) | NC score | 0.612125 (rank : 61) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP28_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 66) | NC score | 0.430122 (rank : 69) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 67) | NC score | 0.689110 (rank : 45) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP29_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 68) | NC score | 0.517302 (rank : 66) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 69) | NC score | 0.517951 (rank : 65) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 70) | NC score | 0.536388 (rank : 63) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
HDAC6_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 71) | NC score | 0.139729 (rank : 76) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
UBP25_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 72) | NC score | 0.407425 (rank : 72) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 73) | NC score | 0.050945 (rank : 85) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
UBP25_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 74) | NC score | 0.404974 (rank : 73) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 75) | NC score | 0.507690 (rank : 67) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 76) | NC score | 0.504618 (rank : 68) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
HDAC6_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 77) | NC score | 0.131038 (rank : 77) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 78) | NC score | 0.049158 (rank : 86) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 79) | NC score | 0.047778 (rank : 87) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
SYT3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 80) | NC score | 0.015022 (rank : 120) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 81) | NC score | 0.046746 (rank : 90) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 82) | NC score | 0.014477 (rank : 122) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 83) | NC score | 0.044511 (rank : 94) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 84) | NC score | 0.016719 (rank : 116) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 85) | NC score | 0.046873 (rank : 89) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 0.21417 (rank : 86) | NC score | 0.421537 (rank : 71) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 87) | NC score | 0.047100 (rank : 88) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 88) | NC score | 0.042467 (rank : 95) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 89) | NC score | 0.056340 (rank : 80) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 90) | NC score | 0.034807 (rank : 97) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TTC3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 91) | NC score | 0.020902 (rank : 109) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 92) | NC score | 0.045651 (rank : 93) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 93) | NC score | 0.012379 (rank : 129) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 94) | NC score | 0.021928 (rank : 108) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 95) | NC score | 0.020457 (rank : 110) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
CIR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 96) | NC score | 0.024565 (rank : 104) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 97) | NC score | 0.046371 (rank : 92) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 98) | NC score | 0.022816 (rank : 106) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SGOL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 99) | NC score | 0.018958 (rank : 114) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
CCD82_HUMAN
|
||||||
| θ value | 1.38821 (rank : 100) | NC score | 0.022608 (rank : 107) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 101) | NC score | 0.029289 (rank : 98) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 1.38821 (rank : 102) | NC score | 0.019840 (rank : 111) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 103) | NC score | -0.000233 (rank : 157) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
DMXL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 104) | NC score | 0.009744 (rank : 136) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 105) | NC score | 0.010648 (rank : 133) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
LIPA3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 106) | NC score | 0.012925 (rank : 126) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
TSP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 107) | NC score | 0.002530 (rank : 153) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
HNRPC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 108) | NC score | 0.013051 (rank : 125) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |
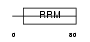
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 109) | NC score | 0.028545 (rank : 100) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 110) | NC score | -0.000005 (rank : 156) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | -0.001809 (rank : 158) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.036968 (rank : 96) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.011582 (rank : 132) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.016528 (rank : 117) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
RIOK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.019507 (rank : 112) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.009844 (rank : 135) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.009083 (rank : 139) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
IL6RB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.004814 (rank : 150) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00560 | Gene names | Il6st | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor beta chain precursor (IL-6R-beta) (Interleukin 6 signal transducer) (Membrane glycoprotein 130) (gp130) (CD130 antigen). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.054113 (rank : 82) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.014977 (rank : 121) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.005814 (rank : 146) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | -0.001933 (rank : 159) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.046527 (rank : 91) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.029211 (rank : 99) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.005060 (rank : 149) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MYG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.027845 (rank : 101) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HB07 | Gene names | C12orf10 | |||
|
Domain Architecture |
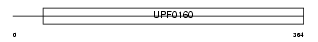
|
|||||
| Description | UPF0160 protein MYG1. | |||||
|
MYG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.027710 (rank : 102) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK81, Q9CYX0 | Gene names | Myg1 | |||
|
Domain Architecture |
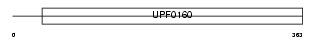
|
|||||
| Description | UPF0160 protein MYG1 (Protein Gamm1). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.014016 (rank : 124) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
NPFF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | -0.005279 (rank : 161) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5X5, Q96RV1, Q9NR49 | Gene names | NPFFR2, GPR74, NPFF2, NPGPR | |||
|
Domain Architecture |

|
|||||
| Description | Neuropeptide FF receptor 2 (Neuropeptide G-protein coupled receptor) (G-protein coupled receptor 74) (G-protein coupled receptor HLWAR77). | |||||
|
PSA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.010127 (rank : 134) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.019317 (rank : 113) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.005783 (rank : 147) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.016521 (rank : 118) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.016976 (rank : 115) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
UBP53_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.054359 (rank : 81) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
APLD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.016414 (rank : 119) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96LR9, Q8IVR2, Q9H0I5 | Gene names | APOLD1, VERGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apolipoprotein-L domain-containing protein 1 (Vascular early response gene protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.003716 (rank : 152) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.007706 (rank : 141) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CF152_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.012567 (rank : 127) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
NFYC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.009373 (rank : 138) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
Domain Architecture |
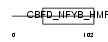
|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | -0.004066 (rank : 160) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.001820 (rank : 154) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TMCO2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.012130 (rank : 131) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z6W1 | Gene names | TMCO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 2. | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.025791 (rank : 103) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ASB6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.001075 (rank : 155) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
BCN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.005626 (rank : 148) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88597 | Gene names | Becn1 | |||
|
Domain Architecture |

|
|||||
| Description | Beclin-1 (Coiled-coil myosin-like BCL2-interacting protein). | |||||
|
CF152_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.014034 (rank : 123) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
CP135_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.003997 (rank : 151) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
ICAL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.009068 (rank : 140) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
JHD2C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.006085 (rank : 145) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.012425 (rank : 128) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.012358 (rank : 130) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.007069 (rank : 142) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PQBP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.023992 (rank : 105) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60828, Q4VY25, Q4VY26, Q4VY27, Q4VY29, Q4VY30, Q4VY34, Q4VY35, Q4VY36, Q4VY37, Q4VY38, Q9GZP2, Q9GZU4, Q9GZZ4 | Gene names | PQBP1, NPW38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
PSA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.009595 (rank : 137) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
SYT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.006683 (rank : 143) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
TRI29_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.006616 (rank : 144) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
GGNB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.075648 (rank : 79) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5YKI7, Q5YKI8 | Gene names | GGNBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
GGNB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.099663 (rank : 78) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6K1E7, Q8C5T9 | Gene names | Ggnbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
UBP54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.052950 (rank : 84) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP54_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.053679 (rank : 83) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 157 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.986072 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.898526 (rank : 3) | θ value | 1.98606e-31 (rank : 7) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.897144 (rank : 4) | θ value | 5.22648e-32 (rank : 3) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.891122 (rank : 5) | θ value | 8.91499e-32 (rank : 4) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.889741 (rank : 6) | θ value | 4.42448e-31 (rank : 8) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.882784 (rank : 7) | θ value | 8.08199e-25 (rank : 17) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.873544 (rank : 8) | θ value | 1.47289e-26 (rank : 11) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.872146 (rank : 9) | θ value | 2.77775e-25 (rank : 16) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.871606 (rank : 10) | θ value | 1.73987e-27 (rank : 9) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.870008 (rank : 11) | θ value | 3.87602e-27 (rank : 10) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.868023 (rank : 12) | θ value | 8.91499e-32 (rank : 5) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.866865 (rank : 13) | θ value | 6.84181e-24 (rank : 21) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.853840 (rank : 14) | θ value | 1.99529e-15 (rank : 35) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.842318 (rank : 15) | θ value | 7.0802e-21 (rank : 24) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.838691 (rank : 16) | θ value | 8.95645e-16 (rank : 32) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.832663 (rank : 17) | θ value | 1.99529e-15 (rank : 36) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.831949 (rank : 18) | θ value | 2.27762e-19 (rank : 26) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.829925 (rank : 19) | θ value | 5.07402e-19 (rank : 27) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.826973 (rank : 20) | θ value | 1.52067e-31 (rank : 6) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.826362 (rank : 21) | θ value | 7.30988e-26 (rank : 14) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.824807 (rank : 22) | θ value | 4.43474e-23 (rank : 22) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.824002 (rank : 23) | θ value | 1.99529e-15 (rank : 34) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.822965 (rank : 24) | θ value | 2.35151e-24 (rank : 18) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.821838 (rank : 25) | θ value | 1.92365e-26 (rank : 13) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.821443 (rank : 26) | θ value | 4.01107e-24 (rank : 20) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.821331 (rank : 27) | θ value | 4.01107e-24 (rank : 19) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.818794 (rank : 28) | θ value | 1.24688e-25 (rank : 15) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.818603 (rank : 29) | θ value | 2.43908e-13 (rank : 41) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.818603 (rank : 30) | θ value | 2.43908e-13 (rank : 42) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.816883 (rank : 31) | θ value | 1.92365e-26 (rank : 12) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.813799 (rank : 32) | θ value | 8.95645e-16 (rank : 33) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.807246 (rank : 33) | θ value | 4.16044e-13 (rank : 46) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.803398 (rank : 34) | θ value | 2.88119e-14 (rank : 37) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.797393 (rank : 35) | θ value | 8.36355e-22 (rank : 23) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.794002 (rank : 36) | θ value | 1.09485e-13 (rank : 38) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.793824 (rank : 37) | θ value | 4.58923e-20 (rank : 25) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.785473 (rank : 38) | θ value | 2.13179e-17 (rank : 29) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.758026 (rank : 39) | θ value | 1.13037e-18 (rank : 28) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.714148 (rank : 40) | θ value | 2.79066e-09 (rank : 54) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.712058 (rank : 41) | θ value | 4.74913e-17 (rank : 30) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.711896 (rank : 42) | θ value | 9.59137e-10 (rank : 53) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.711747 (rank : 43) | θ value | 3.29651e-10 (rank : 52) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.702393 (rank : 44) | θ value | 2.98157e-11 (rank : 51) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.689110 (rank : 45) | θ value | 7.1131e-05 (rank : 67) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.687834 (rank : 46) | θ value | 2.43908e-13 (rank : 40) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.686686 (rank : 47) | θ value | 1.29631e-06 (rank : 58) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.683049 (rank : 48) | θ value | 3.64472e-09 (rank : 55) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.680947 (rank : 49) | θ value | 2.43908e-13 (rank : 39) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.677595 (rank : 50) | θ value | 7.84624e-12 (rank : 49) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.673066 (rank : 51) | θ value | 1.74796e-11 (rank : 50) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.672412 (rank : 52) | θ value | 1.80466e-16 (rank : 31) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.669310 (rank : 53) | θ value | 6.87365e-08 (rank : 56) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.664618 (rank : 54) | θ value | 1.53129e-07 (rank : 57) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.659749 (rank : 55) | θ value | 4.16044e-13 (rank : 47) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.659234 (rank : 56) | θ value | 3.18553e-13 (rank : 45) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.646717 (rank : 57) | θ value | 2.43908e-13 (rank : 43) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.646166 (rank : 58) | θ value | 2.43908e-13 (rank : 44) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.644711 (rank : 59) | θ value | 1.2105e-12 (rank : 48) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.621194 (rank : 60) | θ value | 1.87187e-05 (rank : 62) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.612125 (rank : 61) | θ value | 2.44474e-05 (rank : 65) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP14_MOUSE
|
||||||
| NC score | 0.607134 (rank : 62) | θ value | 2.44474e-05 (rank : 64) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.536388 (rank : 63) | θ value | 0.00035302 (rank : 70) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.519937 (rank : 64) | θ value | 2.21117e-06 (rank : 59) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.517951 (rank : 65) | θ value | 0.00035302 (rank : 69) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP29_MOUSE
|
||||||
| NC score | 0.517302 (rank : 66) | θ value | 9.29e-05 (rank : 68) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.507690 (rank : 67) | θ value | 0.00134147 (rank : 75) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.504618 (rank : 68) | θ value | 0.00134147 (rank : 76) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP28_HUMAN
|
||||||
| NC score | 0.430122 (rank : 69) | θ value | 7.1131e-05 (rank : 66) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.427765 (rank : 70) | θ value | 1.87187e-05 (rank : 63) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.421537 (rank : 71) | θ value | 0.21417 (rank : 86) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP25_MOUSE
|
||||||
| NC score | 0.407425 (rank : 72) | θ value | 0.000602161 (rank : 72) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP25_HUMAN
|
||||||
| NC score | 0.404974 (rank : 73) | θ value | 0.000786445 (rank : 74) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
BRAP_MOUSE
|
||||||
| NC score | 0.212450 (rank : 74) | θ value | 6.43352e-06 (rank : 61) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
BRAP_HUMAN
|
||||||
| NC score | 0.188087 (rank : 75) | θ value | 6.43352e-06 (rank : 60) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
HDAC6_MOUSE
|
||||||
| NC score | 0.139729 (rank : 76) | θ value | 0.000461057 (rank : 71) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
HDAC6_HUMAN
|
||||||
| NC score | 0.131038 (rank : 77) | θ value | 0.00298849 (rank : 77) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
GGNB1_MOUSE
|
||||||
| NC score | 0.099663 (rank : 78) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6K1E7, Q8C5T9 | Gene names | Ggnbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
GGNB1_HUMAN
|
||||||
| NC score | 0.075648 (rank : 79) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5YKI7, Q5YKI8 | Gene names | GGNBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.056340 (rank : 80) | θ value | 0.279714 (rank : 89) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
UBP53_HUMAN
|
||||||
| NC score | 0.054359 (rank : 81) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.054113 (rank : 82) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
UBP54_MOUSE
|
||||||
| NC score | 0.053679 (rank : 83) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP54_HUMAN
|
||||||
| NC score | 0.052950 (rank : 84) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.050945 (rank : 85) | θ value | 0.000786445 (rank : 73) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.049158 (rank : 86) | θ value | 0.00298849 (rank : 78) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.047778 (rank : 87) | θ value | 0.00390308 (rank : 79) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.047100 (rank : 88) | θ value | 0.279714 (rank : 87) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.046873 (rank : 89) | θ value | 0.21417 (rank : 85) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.046746 (rank : 90) | θ value | 0.0330416 (rank : 81) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.046527 (rank : 91) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.046371 (rank : 92) | θ value | 1.06291 (rank : 97) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.045651 (rank : 93) | θ value | 0.47712 (rank : 92) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.044511 (rank : 94) | θ value | 0.0736092 (rank : 83) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.042467 (rank : 95) | θ value | 0.279714 (rank : 88) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.036968 (rank : 96) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.034807 (rank : 97) | θ value | 0.279714 (rank : 90) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.029289 (rank : 98) | θ value | 1.38821 (rank : 101) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.029211 (rank : 99) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.028545 (rank : 100) | θ value | 2.36792 (rank : 109) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MYG1_HUMAN
|
||||||
| NC score | 0.027845 (rank : 101) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HB07 | Gene names | C12orf10 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0160 protein MYG1. | |||||
|
MYG1_MOUSE
|
||||||
| NC score | 0.027710 (rank : 102) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK81, Q9CYX0 | Gene names | Myg1 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0160 protein MYG1 (Protein Gamm1). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.025791 (rank : 103) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.024565 (rank : 104) | θ value | 1.06291 (rank : 96) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
PQBP1_HUMAN
|
||||||
| NC score | 0.023992 (rank : 105) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60828, Q4VY25, Q4VY26, Q4VY27, Q4VY29, Q4VY30, Q4VY34, Q4VY35, Q4VY36, Q4VY37, Q4VY38, Q9GZP2, Q9GZU4, Q9GZZ4 | Gene names | PQBP1, NPW38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyglutamine-binding protein 1 (Polyglutamine tract-binding protein 1) (PQBP-1) (38 kDa nuclear protein containing a WW domain) (Npw38). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.022816 (rank : 106) | θ value | 1.06291 (rank : 98) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CCD82_HUMAN
|
||||||
| NC score | 0.022608 (rank : 107) | θ value | 1.38821 (rank : 100) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.021928 (rank : 108) | θ value | 0.62314 (rank : 94) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.020902 (rank : 109) | θ value | 0.365318 (rank : 91) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.020457 (rank : 110) | θ value | 0.813845 (rank : 95) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.019840 (rank : 111) | θ value | 1.38821 (rank : 102) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
RIOK1_HUMAN
|
||||||
| NC score | 0.019507 (rank : 112) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.019317 (rank : 113) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SGOL2_HUMAN
|
||||||
| NC score | 0.018958 (rank : 114) | θ value | 1.06291 (rank : 99) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q562F6, Q53RR9, Q53T20, Q86XY4, Q8IWK2, Q8IZK1, Q8N1Q5, Q96LQ3 | Gene names | SGOL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2 (Tripin). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.016976 (rank : 115) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.016719 (rank : 116) | θ value | 0.21417 (rank : 84) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.016528 (rank : 117) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.016521 (rank : 118) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
APLD1_HUMAN
|
||||||
| NC score | 0.016414 (rank : 119) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96LR9, Q8IVR2, Q9H0I5 | Gene names | APOLD1, VERGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apolipoprotein-L domain-containing protein 1 (Vascular early response gene protein). | |||||
|
SYT3_MOUSE
|
||||||
| NC score | 0.015022 (rank : 120) | θ value | 0.0193708 (rank : 80) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.014977 (rank : 121) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.014477 (rank : 122) | θ value | 0.0736092 (rank : 82) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.014034 (rank : 123) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.014016 (rank : 124) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
HNRPC_HUMAN
|
||||||
| NC score | 0.013051 (rank : 125) | θ value | 2.36792 (rank : 108) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
LIPA3_MOUSE
|
||||||
| NC score | 0.012925 (rank : 126) | θ value | 1.81305 (rank : 106) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.012567 (rank : 127) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.012425 (rank : 128) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.012379 (rank : 129) | θ value | 0.47712 (rank : 93) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.012358 (rank : 130) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
TMCO2_HUMAN
|
||||||
| NC score | 0.012130 (rank : 131) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z6W1 | Gene names | TMCO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 2. | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.011582 (rank : 132) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.010648 (rank : 133) | θ value | 1.81305 (rank : 105) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
PSA_HUMAN
|
||||||
| NC score | 0.010127 (rank : 134) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.009844 (rank : 135) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
DMXL1_HUMAN
|
||||||
| NC score | 0.009744 (rank : 136) | θ value | 1.81305 (rank : 104) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
PSA_MOUSE
|
||||||
| NC score | 0.009595 (rank : 137) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
NFYC_HUMAN
|
||||||
| NC score | 0.009373 (rank : 138) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.009083 (rank : 139) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.009068 (rank : 140) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.007706 (rank : 141) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.007069 (rank : 142) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
SYT3_HUMAN
|
||||||
| NC score | 0.006683 (rank : 143) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
TRI29_HUMAN
|
||||||
| NC score | 0.006616 (rank : 144) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
JHD2C_HUMAN
|
||||||
| NC score | 0.006085 (rank : 145) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.005814 (rank : 146) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.005783 (rank : 147) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
BCN1_MOUSE
|
||||||
| NC score | 0.005626 (rank : 148) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88597 | Gene names | Becn1 | |||
|
Domain Architecture |

|
|||||
| Description | Beclin-1 (Coiled-coil myosin-like BCL2-interacting protein). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.005060 (rank : 149) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
IL6RB_MOUSE
|
||||||
| NC score | 0.004814 (rank : 150) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00560 | Gene names | Il6st | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor beta chain precursor (IL-6R-beta) (Interleukin 6 signal transducer) (Membrane glycoprotein 130) (gp130) (CD130 antigen). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.003997 (rank : 151) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.003716 (rank : 152) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
TSP2_MOUSE
|
||||||
| NC score | 0.002530 (rank : 153) | θ value | 1.81305 (rank : 107) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.001820 (rank : 154) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ASB6_MOUSE
|
||||||
| NC score | 0.001075 (rank : 155) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | -0.000005 (rank : 156) | θ value | 2.36792 (rank : 110) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | -0.000233 (rank : 157) | θ value | 1.81305 (rank : 103) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | -0.001809 (rank : 158) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | -0.001933 (rank : 159) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
SLK_HUMAN
|
||||||
| NC score | -0.004066 (rank : 160) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
NPFF2_HUMAN
|
||||||
| NC score | -0.005279 (rank : 161) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5X5, Q96RV1, Q9NR49 | Gene names | NPFFR2, GPR74, NPFF2, NPGPR | |||
|
Domain Architecture |

|
|||||
| Description | Neuropeptide FF receptor 2 (Neuropeptide G-protein coupled receptor) (G-protein coupled receptor 74) (G-protein coupled receptor HLWAR77). | |||||