Please be patient as the page loads
|
UBP20_HUMAN
|
||||||
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBP20_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.986072 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 3) | NC score | 0.901496 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 2.19584e-30 (rank : 4) | NC score | 0.901553 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 2.86786e-30 (rank : 5) | NC score | 0.868459 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 6) | NC score | 0.826441 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 1.85889e-29 (rank : 7) | NC score | 0.892466 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 8) | NC score | 0.891838 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 9) | NC score | 0.877146 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 10) | NC score | 0.875667 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 11) | NC score | 0.819789 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 12) | NC score | 0.873378 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 13) | NC score | 0.829121 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 14) | NC score | 0.820770 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 15) | NC score | 0.824524 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 16) | NC score | 0.806222 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 17) | NC score | 0.875259 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 18) | NC score | 0.883359 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 19) | NC score | 0.873536 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 1.5242e-23 (rank : 20) | NC score | 0.802601 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 21) | NC score | 0.816891 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 22) | NC score | 0.826040 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 23) | NC score | 0.820025 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 24) | NC score | 0.820276 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 6.40375e-22 (rank : 25) | NC score | 0.853492 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 26) | NC score | 0.842635 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 27) | NC score | 0.840612 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 28) | NC score | 0.760363 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 29) | NC score | 0.672407 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 30) | NC score | 0.712382 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 31) | NC score | 0.823562 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 32) | NC score | 0.809572 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 33) | NC score | 0.837038 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 34) | NC score | 0.788630 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 35) | NC score | 0.837957 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 36) | NC score | 0.804542 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 37) | NC score | 0.709469 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 38) | NC score | 0.857685 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 39) | NC score | 0.826361 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 40) | NC score | 0.653330 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 41) | NC score | 0.662930 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 42) | NC score | 0.820748 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 43) | NC score | 0.820748 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 44) | NC score | 0.652802 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 2.0648e-12 (rank : 45) | NC score | 0.662930 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 46) | NC score | 0.652127 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UB17L_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 47) | NC score | 0.811726 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 48) | NC score | 0.670914 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 49) | NC score | 0.677747 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 50) | NC score | 0.724051 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 51) | NC score | 0.721109 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 52) | NC score | 0.669920 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 53) | NC score | 0.704145 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 54) | NC score | 0.727231 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 55) | NC score | 0.665952 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 56) | NC score | 0.693866 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
HDAC6_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 57) | NC score | 0.154897 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 58) | NC score | 0.655527 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 59) | NC score | 0.661555 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 60) | NC score | 0.613329 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
HDAC6_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 61) | NC score | 0.145716 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 62) | NC score | 0.512367 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
BRAP_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 63) | NC score | 0.186440 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
BRAP_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 64) | NC score | 0.212177 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 65) | NC score | 0.540635 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 66) | NC score | 0.618850 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP14_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 67) | NC score | 0.603002 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 68) | NC score | 0.520456 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP29_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 69) | NC score | 0.520599 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 70) | NC score | 0.498893 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 71) | NC score | 0.682533 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 72) | NC score | 0.501529 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP28_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 73) | NC score | 0.403198 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 74) | NC score | 0.425987 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 75) | NC score | 0.401037 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP25_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 76) | NC score | 0.381186 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP25_HUMAN
|
||||||
| θ value | 0.125558 (rank : 77) | NC score | 0.378515 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
ZHX2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 78) | NC score | 0.022195 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6X8 | Gene names | ZHX2, AFR1, KIAA0854, RAF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
ZHX2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.019701 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.025609 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MYG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.037232 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HB07 | Gene names | C12orf10 | |||
|
Domain Architecture |
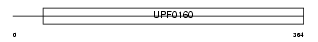
|
|||||
| Description | UPF0160 protein MYG1. | |||||
|
MYG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.036944 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK81, Q9CYX0 | Gene names | Myg1 | |||
|
Domain Architecture |
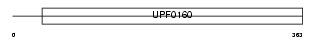
|
|||||
| Description | UPF0160 protein MYG1 (Protein Gamm1). | |||||
|
SCRT2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | -0.003341 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ03 | Gene names | SCRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor scratch 2 (Scratch homolog 2 zinc finger protein). | |||||
|
ZFP60_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | -0.003882 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16374, Q61135 | Gene names | Zfp60, Mfg3 | |||
|
Domain Architecture |
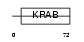
|
|||||
| Description | Zinc finger protein 60 (Zfp-60) (Zinc finger protein Mfg-3). | |||||
|
PSA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.014194 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
ZN426_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | -0.004531 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BUY5 | Gene names | ZNF426 | |||
|
Domain Architecture |
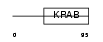
|
|||||
| Description | Zinc finger protein 426. | |||||
|
APXL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.011637 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
CJ119_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.016154 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTE3, Q6IA56, Q9BVT9, Q9H916 | Gene names | C10orf119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119. | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.008494 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
LOXL2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.009329 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
PSA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.014551 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.024200 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LOXL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.008920 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
STK4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | -0.006324 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13043, Q15802, Q4G156, Q5H982, Q6PD60, Q9BR32, Q9NTZ4 | Gene names | STK4, MST1 | |||
|
Domain Architecture |
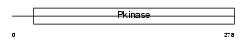
|
|||||
| Description | Serine/threonine-protein kinase 4 (EC 2.7.11.1) (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1) (Serine/threonine-protein kinase Krs-2). | |||||
|
URFB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.017861 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6BDS2, Q9NXE0 | Gene names | UHRF1BP1, C6orf107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UHRF1-binding protein 1 (Ubiquitin-like containing PHD and RING finger domains 1-binding protein 1) (ICBP90-binding protein 1). | |||||
|
HDGF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.011162 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.016590 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
NTNG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.003880 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |
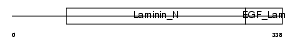
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
XPO6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.017080 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q924Z6, Q3U3K7, Q6PEF3, Q9D0K9 | Gene names | Xpo6, Ranbp20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-6 (Exp6) (Ran-binding protein 20). | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.007923 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
SSR3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | -0.004408 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32745 | Gene names | SSTR3 | |||
|
Domain Architecture |
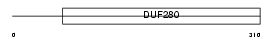
|
|||||
| Description | Somatostatin receptor type 3 (SS3R) (SSR-28). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.002630 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
XPO6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.016315 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QU8, Q2YDX3, Q53G88, Q68G50, Q76N88, Q96CP8, Q9BT21 | Gene names | XPO6, KIAA0370, RANBP20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-6 (Exp6) (Ran-binding protein 20). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.001201 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | -0.001256 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
DCDC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.006365 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
NTNG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.003144 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |
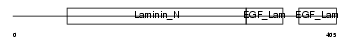
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
PP2BB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.004699 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16298, P16299, Q8N3W4 | Gene names | PPP3CB, CALNA2, CNA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit beta isoform) (CAM-PRP catalytic subunit). | |||||
|
PP2BB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.004705 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48453, Q6NZR4 | Gene names | Ppp3cb, Calnb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit beta isoform) (CAM-PRP catalytic subunit). | |||||
|
F263_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.004920 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16875, O43622, O75902 | Gene names | PFKFB3 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (6PF-2-K/Fru- 2,6-P2ASE brain/placenta-type isozyme) (iPFK-2) (NY-REN-56 antigen) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6- bisphosphatase (EC 3.1.3.46)]. | |||||
|
SYT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.007503 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.018715 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
VINEX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.001180 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
GGNB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.074074 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5YKI7, Q5YKI8 | Gene names | GGNBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
GGNB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.097668 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6K1E7, Q8C5T9 | Gene names | Ggnbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
TBCD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.051530 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8IZP1, Q9H0B9, Q9UDD4 | Gene names | TBC1D3, PRC17 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 3 (Rab GTPase-activating protein PRC17) (Prostate cancer gene 17 protein) (TRE17 alpha protein). | |||||
|
UBP54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.051106 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP54_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.051845 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.986072 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.901553 (rank : 3) | θ value | 2.19584e-30 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.901496 (rank : 4) | θ value | 2.19584e-30 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.892466 (rank : 5) | θ value | 1.85889e-29 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.891838 (rank : 6) | θ value | 1.57365e-28 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.883359 (rank : 7) | θ value | 6.84181e-24 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.877146 (rank : 8) | θ value | 1.73987e-27 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.875667 (rank : 9) | θ value | 1.73987e-27 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.875259 (rank : 10) | θ value | 6.84181e-24 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.873536 (rank : 11) | θ value | 1.5242e-23 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.873378 (rank : 12) | θ value | 1.24688e-25 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.868459 (rank : 13) | θ value | 2.86786e-30 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.857685 (rank : 14) | θ value | 3.76295e-14 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.853492 (rank : 15) | θ value | 6.40375e-22 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.842635 (rank : 16) | θ value | 8.36355e-22 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.840612 (rank : 17) | θ value | 1.42661e-21 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.837957 (rank : 18) | θ value | 5.8054e-15 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.837038 (rank : 19) | θ value | 1.52774e-15 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.829121 (rank : 20) | θ value | 3.62785e-25 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.826441 (rank : 21) | θ value | 4.89182e-30 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.826361 (rank : 22) | θ value | 1.42992e-13 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.826040 (rank : 23) | θ value | 9.87957e-23 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.824524 (rank : 24) | θ value | 5.23862e-24 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.823562 (rank : 25) | θ value | 6.20254e-17 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.820770 (rank : 26) | θ value | 8.08199e-25 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.820748 (rank : 27) | θ value | 7.09661e-13 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.820748 (rank : 28) | θ value | 7.09661e-13 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.820276 (rank : 29) | θ value | 2.87452e-22 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.820025 (rank : 30) | θ value | 2.87452e-22 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.819789 (rank : 31) | θ value | 7.30988e-26 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.816891 (rank : 32) | θ value | 2.59989e-23 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.811726 (rank : 33) | θ value | 1.02475e-11 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.809572 (rank : 34) | θ value | 8.95645e-16 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.806222 (rank : 35) | θ value | 5.23862e-24 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.804542 (rank : 36) | θ value | 2.88119e-14 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.802601 (rank : 37) | θ value | 1.5242e-23 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.788630 (rank : 38) | θ value | 2.60593e-15 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.760363 (rank : 39) | θ value | 2.5182e-18 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.727231 (rank : 40) | θ value | 3.29651e-10 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.724051 (rank : 41) | θ value | 5.08577e-11 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.721109 (rank : 42) | θ value | 6.64225e-11 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.712382 (rank : 43) | θ value | 4.74913e-17 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.709469 (rank : 44) | θ value | 2.88119e-14 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.704145 (rank : 45) | θ value | 6.64225e-11 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.693866 (rank : 46) | θ value | 6.87365e-08 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.682533 (rank : 47) | θ value | 0.0113563 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.677747 (rank : 48) | θ value | 2.98157e-11 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.672407 (rank : 49) | θ value | 3.63628e-17 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.670914 (rank : 50) | θ value | 2.98157e-11 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.669920 (rank : 51) | θ value | 6.64225e-11 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.665952 (rank : 52) | θ value | 4.30538e-10 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.662930 (rank : 53) | θ value | 7.09661e-13 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.662930 (rank : 54) | θ value | 2.0648e-12 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.661555 (rank : 55) | θ value | 6.43352e-06 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.655527 (rank : 56) | θ value | 6.43352e-06 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.653330 (rank : 57) | θ value | 2.43908e-13 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.652802 (rank : 58) | θ value | 7.09661e-13 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.652127 (rank : 59) | θ value | 2.0648e-12 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.618850 (rank : 60) | θ value | 0.000158464 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.613329 (rank : 61) | θ value | 1.09739e-05 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP14_MOUSE
|
||||||
| NC score | 0.603002 (rank : 62) | θ value | 0.000158464 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.540635 (rank : 63) | θ value | 4.1701e-05 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP29_MOUSE
|
||||||
| NC score | 0.520599 (rank : 64) | θ value | 0.00020696 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.520456 (rank : 65) | θ value | 0.00020696 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.512367 (rank : 66) | θ value | 2.44474e-05 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.501529 (rank : 67) | θ value | 0.0148317 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.498893 (rank : 68) | θ value | 0.00869519 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.425987 (rank : 69) | θ value | 0.0252991 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP28_HUMAN
|
||||||
| NC score | 0.403198 (rank : 70) | θ value | 0.0193708 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.401037 (rank : 71) | θ value | 0.0431538 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP25_MOUSE
|
||||||
| NC score | 0.381186 (rank : 72) | θ value | 0.0736092 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP25_HUMAN
|
||||||
| NC score | 0.378515 (rank : 73) | θ value | 0.125558 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
BRAP_MOUSE
|
||||||
| NC score | 0.212177 (rank : 74) | θ value | 3.19293e-05 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
BRAP_HUMAN
|
||||||
| NC score | 0.186440 (rank : 75) | θ value | 3.19293e-05 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
HDAC6_MOUSE
|
||||||
| NC score | 0.154897 (rank : 76) | θ value | 4.92598e-06 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
HDAC6_HUMAN
|
||||||
| NC score | 0.145716 (rank : 77) | θ value | 2.44474e-05 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
GGNB1_MOUSE
|
||||||
| NC score | 0.097668 (rank : 78) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6K1E7, Q8C5T9 | Gene names | Ggnbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
GGNB1_HUMAN
|
||||||
| NC score | 0.074074 (rank : 79) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5YKI7, Q5YKI8 | Gene names | GGNBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
UBP54_MOUSE
|
||||||
| NC score | 0.051845 (rank : 80) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
TBCD3_HUMAN
|
||||||
| NC score | 0.051530 (rank : 81) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q8IZP1, Q9H0B9, Q9UDD4 | Gene names | TBC1D3, PRC17 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 3 (Rab GTPase-activating protein PRC17) (Prostate cancer gene 17 protein) (TRE17 alpha protein). | |||||
|
UBP54_HUMAN
|
||||||
| NC score | 0.051106 (rank : 82) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
MYG1_HUMAN
|
||||||
| NC score | 0.037232 (rank : 83) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HB07 | Gene names | C12orf10 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0160 protein MYG1. | |||||
|
MYG1_MOUSE
|
||||||
| NC score | 0.036944 (rank : 84) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JK81, Q9CYX0 | Gene names | Myg1 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0160 protein MYG1 (Protein Gamm1). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.025609 (rank : 85) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.024200 (rank : 86) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
ZHX2_HUMAN
|
||||||
| NC score | 0.022195 (rank : 87) | θ value | 0.365318 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6X8 | Gene names | ZHX2, AFR1, KIAA0854, RAF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
ZHX2_MOUSE
|
||||||
| NC score | 0.019701 (rank : 88) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.018715 (rank : 89) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
URFB1_HUMAN
|
||||||
| NC score | 0.017861 (rank : 90) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6BDS2, Q9NXE0 | Gene names | UHRF1BP1, C6orf107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UHRF1-binding protein 1 (Ubiquitin-like containing PHD and RING finger domains 1-binding protein 1) (ICBP90-binding protein 1). | |||||
|
XPO6_MOUSE
|
||||||
| NC score | 0.017080 (rank : 91) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q924Z6, Q3U3K7, Q6PEF3, Q9D0K9 | Gene names | Xpo6, Ranbp20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-6 (Exp6) (Ran-binding protein 20). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.016590 (rank : 92) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
XPO6_HUMAN
|
||||||
| NC score | 0.016315 (rank : 93) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QU8, Q2YDX3, Q53G88, Q68G50, Q76N88, Q96CP8, Q9BT21 | Gene names | XPO6, KIAA0370, RANBP20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-6 (Exp6) (Ran-binding protein 20). | |||||
|
CJ119_HUMAN
|
||||||
| NC score | 0.016154 (rank : 94) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTE3, Q6IA56, Q9BVT9, Q9H916 | Gene names | C10orf119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf119. | |||||
|
PSA_HUMAN
|
||||||
| NC score | 0.014551 (rank : 95) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55786, Q6P145, Q9NP16, Q9UEM2 | Gene names | NPEPPS, PSA | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
PSA_MOUSE
|
||||||
| NC score | 0.014194 (rank : 96) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q11011, Q91VJ8 | Gene names | Npepps, Psa | |||
|
Domain Architecture |

|
|||||
| Description | Puromycin-sensitive aminopeptidase (EC 3.4.11.-) (PSA). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.011637 (rank : 97) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
HDGF_MOUSE
|
||||||
| NC score | 0.011162 (rank : 98) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
LOXL2_MOUSE
|
||||||
| NC score | 0.009329 (rank : 99) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
LOXL2_HUMAN
|
||||||
| NC score | 0.008920 (rank : 100) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.008494 (rank : 101) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.007923 (rank : 102) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
SYT3_MOUSE
|
||||||
| NC score | 0.007503 (rank : 103) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
DCDC2_MOUSE
|
||||||
| NC score | 0.006365 (rank : 104) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
F263_HUMAN
|
||||||
| NC score | 0.004920 (rank : 105) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16875, O43622, O75902 | Gene names | PFKFB3 | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (6PF-2-K/Fru- 2,6-P2ASE brain/placenta-type isozyme) (iPFK-2) (NY-REN-56 antigen) [Includes: 6-phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6- bisphosphatase (EC 3.1.3.46)]. | |||||
|
PP2BB_MOUSE
|
||||||
| NC score | 0.004705 (rank : 106) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48453, Q6NZR4 | Gene names | Ppp3cb, Calnb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit beta isoform) (CAM-PRP catalytic subunit). | |||||
|
PP2BB_HUMAN
|
||||||
| NC score | 0.004699 (rank : 107) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16298, P16299, Q8N3W4 | Gene names | PPP3CB, CALNA2, CNA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (EC 3.1.3.16) (Calmodulin-dependent calcineurin A subunit beta isoform) (CAM-PRP catalytic subunit). | |||||
|
NTNG1_HUMAN
|
||||||
| NC score | 0.003880 (rank : 108) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG1_MOUSE
|
||||||
| NC score | 0.003144 (rank : 109) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.002630 (rank : 110) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.001201 (rank : 111) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
VINEX_MOUSE
|
||||||
| NC score | 0.001180 (rank : 112) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | -0.001256 (rank : 113) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
SCRT2_HUMAN
|
||||||
| NC score | -0.003341 (rank : 114) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ03 | Gene names | SCRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor scratch 2 (Scratch homolog 2 zinc finger protein). | |||||
|
ZFP60_MOUSE
|
||||||
| NC score | -0.003882 (rank : 115) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16374, Q61135 | Gene names | Zfp60, Mfg3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 60 (Zfp-60) (Zinc finger protein Mfg-3). | |||||
|
SSR3_HUMAN
|
||||||
| NC score | -0.004408 (rank : 116) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32745 | Gene names | SSTR3 | |||
|
Domain Architecture |

|
|||||
| Description | Somatostatin receptor type 3 (SS3R) (SSR-28). | |||||
|
ZN426_HUMAN
|
||||||
| NC score | -0.004531 (rank : 117) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BUY5 | Gene names | ZNF426 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 426. | |||||
|
STK4_HUMAN
|
||||||
| NC score | -0.006324 (rank : 118) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13043, Q15802, Q4G156, Q5H982, Q6PD60, Q9BR32, Q9NTZ4 | Gene names | STK4, MST1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 4 (EC 2.7.11.1) (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1) (Serine/threonine-protein kinase Krs-2). | |||||