Please be patient as the page loads
|
UBP48_MOUSE
|
||||||
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBP48_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997827 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 1.20211e-36 (rank : 3) | NC score | 0.880344 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 8.61488e-35 (rank : 4) | NC score | 0.796683 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 8.61488e-35 (rank : 5) | NC score | 0.797785 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 6) | NC score | 0.779538 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 7) | NC score | 0.838589 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 8) | NC score | 0.832109 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 9) | NC score | 0.835126 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 10) | NC score | 0.775505 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 11) | NC score | 0.772566 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 12) | NC score | 0.853670 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | 2.35151e-24 (rank : 13) | NC score | 0.853670 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 14) | NC score | 0.786104 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 15) | NC score | 0.801047 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 16) | NC score | 0.746846 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP20_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 17) | NC score | 0.802601 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 18) | NC score | 0.786913 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 19) | NC score | 0.841510 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 20) | NC score | 0.848376 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 21) | NC score | 0.854886 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 3.17815e-21 (rank : 22) | NC score | 0.829608 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UB17L_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 23) | NC score | 0.817731 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 24) | NC score | 0.799577 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 25) | NC score | 0.810273 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 26) | NC score | 0.831795 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 27) | NC score | 0.793824 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 28) | NC score | 0.801354 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 29) | NC score | 0.777151 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 30) | NC score | 0.777303 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 31) | NC score | 0.774616 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP25_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 32) | NC score | 0.581865 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP25_MOUSE
|
||||||
| θ value | 7.32683e-18 (rank : 33) | NC score | 0.583221 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 34) | NC score | 0.796224 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 35) | NC score | 0.780714 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 36) | NC score | 0.790194 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 37) | NC score | 0.780741 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 38) | NC score | 0.731804 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 39) | NC score | 0.794214 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 40) | NC score | 0.794521 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 41) | NC score | 0.794075 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP28_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 42) | NC score | 0.589569 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 43) | NC score | 0.583392 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 44) | NC score | 0.692885 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 45) | NC score | 0.692549 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 46) | NC score | 0.683411 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 47) | NC score | 0.682501 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 48) | NC score | 0.690530 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 49) | NC score | 0.688163 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 50) | NC score | 0.685486 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 51) | NC score | 0.760724 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 52) | NC score | 0.686750 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 53) | NC score | 0.708400 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 54) | NC score | 0.673915 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 55) | NC score | 0.643215 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 56) | NC score | 0.645152 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 57) | NC score | 0.665972 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 58) | NC score | 0.638492 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 59) | NC score | 0.722098 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP14_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 60) | NC score | 0.612226 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 61) | NC score | 0.645902 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 62) | NC score | 0.480188 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 63) | NC score | 0.490359 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 64) | NC score | 0.474780 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 65) | NC score | 0.474383 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 66) | NC score | 0.567420 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 67) | NC score | 0.548285 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 68) | NC score | 0.517281 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP29_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 69) | NC score | 0.464360 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
CALX_HUMAN
|
||||||
| θ value | 0.21417 (rank : 70) | NC score | 0.036551 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
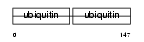
UCRP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 71) | NC score | 0.073435 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64339, Q8K0H3 | Gene names | Isg15, G1p2, Ucrp | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin cross-reactive protein precursor (Interferon-stimulated protein 15) (IP17). | |||||
|
DGKH_HUMAN
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.019220 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.445758 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
CAN3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 74) | NC score | 0.008229 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.015773 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
ZRAB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 76) | NC score | 0.054260 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
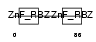
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
ARS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.033640 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 78) | NC score | 0.009141 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
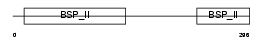
SIAL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.034268 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |
|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
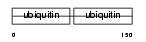
UCRP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 80) | NC score | 0.055997 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05161, Q7Z2G2, Q96GF0 | Gene names | ISG15, G1P2, UCRP | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-induced 17 kDa protein precursor [Contains: Ubiquitin cross-reactive protein (hUCRP) (Interferon-induced 15 kDa protein)]. | |||||
|
PAR12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.013428 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.005375 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.482722 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.494594 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
CSTN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.006870 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94985 | Gene names | CLSTN1, CS1, KIAA0911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.021281 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.004351 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RNF8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.007006 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76064 | Gene names | RNF8, KIAA0646 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8). | |||||
|
STAC3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.015584 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BZ71 | Gene names | Stac3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.012997 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
TF3C5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.018435 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Q8, Q8N2U7, Q8N857, Q96GD9, Q9H4P2 | Gene names | GTF3C5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 5 (Transcription factor IIIC-epsilon subunit) (TF3C-epsilon) (TFIIIC 63 kDa subunit) (TFIIIC63). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.013220 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
FBXW7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.004378 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.010253 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NO40_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.015196 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESX4 | Gene names | Zcchc17, Ps1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein). | |||||
|
ALBU_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.010165 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02768, O95574, P04277, Q13140, Q68DN5, Q6UXK4, Q9P157, Q9P1I7, Q9UHS3, Q9UJZ0 | Gene names | ALB | |||
|
Domain Architecture |

|
|||||
| Description | Serum albumin precursor. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | -0.000704 (rank : 128) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ARS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.024429 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.018304 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
DEK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.016962 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DEK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.021413 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.000281 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.007847 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.373467 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.005388 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
AKP8L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.007329 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
GDAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.010677 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88741, Q8C7Q5, Q9CTN2 | Gene names | Gdap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
MINK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | -0.005861 (rank : 130) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.008751 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.013489 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PSD7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.008263 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26516, Q3TW61, Q8C0I8 | Gene names | Psmd7, Mov-34, Mov34 | |||
|
Domain Architecture |
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.013203 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.015791 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.004222 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | -0.000946 (rank : 129) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BRWD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | -0.000067 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.018940 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
HDGF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.008585 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
KPCT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | -0.006034 (rank : 132) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02111 | Gene names | Prkcq, Pkcq | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
MKNK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | -0.005909 (rank : 131) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDB0, O08606, Q75PY0, Q9D893 | Gene names | Mknk2, Mnk2 | |||
|
Domain Architecture |
|
|||||
| Description | MAP kinase-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (MAP kinase signal-integrating kinase 2) (Mnk2). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.018369 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.006166 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
TF3C5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.014334 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2T8, Q8BLE3, Q8R0D0 | Gene names | Gtf3c5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 5 (Transcription factor IIIC-epsilon subunit) (TF3C-epsilon) (TFIIIC 63 kDa subunit) (TFIIIC63). | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.003213 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
BRAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.084778 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
BRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.095952 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
GGNB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.052532 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5YKI7, Q5YKI8 | Gene names | GGNBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
GGNB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.067661 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6K1E7, Q8C5T9 | Gene names | Ggnbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
HDAC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.063102 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
HDAC6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.065237 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
UBP54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.052200 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP54_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052354 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 124 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.997827 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.880344 (rank : 3) | θ value | 1.20211e-36 (rank : 3) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.854886 (rank : 4) | θ value | 2.43343e-21 (rank : 21) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.853670 (rank : 5) | θ value | 2.35151e-24 (rank : 12) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.853670 (rank : 6) | θ value | 2.35151e-24 (rank : 13) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.848376 (rank : 7) | θ value | 1.6852e-22 (rank : 20) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.841510 (rank : 8) | θ value | 5.79196e-23 (rank : 19) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.838589 (rank : 9) | θ value | 1.0531e-32 (rank : 7) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.835126 (rank : 10) | θ value | 5.77852e-31 (rank : 9) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.832109 (rank : 11) | θ value | 1.52067e-31 (rank : 8) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.831795 (rank : 12) | θ value | 4.58923e-20 (rank : 26) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.829608 (rank : 13) | θ value | 3.17815e-21 (rank : 22) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.817731 (rank : 14) | θ value | 4.15078e-21 (rank : 23) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.810273 (rank : 15) | θ value | 2.69047e-20 (rank : 25) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.802601 (rank : 16) | θ value | 1.5242e-23 (rank : 17) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.801354 (rank : 17) | θ value | 1.74391e-19 (rank : 28) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.801047 (rank : 18) | θ value | 8.93572e-24 (rank : 15) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.799577 (rank : 19) | θ value | 5.42112e-21 (rank : 24) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.797785 (rank : 20) | θ value | 8.61488e-35 (rank : 5) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.796683 (rank : 21) | θ value | 8.61488e-35 (rank : 4) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.796224 (rank : 22) | θ value | 7.32683e-18 (rank : 34) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.794521 (rank : 23) | θ value | 2.60593e-15 (rank : 40) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.794214 (rank : 24) | θ value | 1.99529e-15 (rank : 39) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.794075 (rank : 25) | θ value | 2.60593e-15 (rank : 41) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.793824 (rank : 26) | θ value | 4.58923e-20 (rank : 27) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.790194 (rank : 27) | θ value | 1.058e-16 (rank : 36) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.786913 (rank : 28) | θ value | 4.43474e-23 (rank : 18) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.786104 (rank : 29) | θ value | 4.01107e-24 (rank : 14) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.780741 (rank : 30) | θ value | 1.80466e-16 (rank : 37) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.780714 (rank : 31) | θ value | 1.058e-16 (rank : 35) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.779538 (rank : 32) | θ value | 4.72714e-33 (rank : 6) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.777303 (rank : 33) | θ value | 3.88503e-19 (rank : 30) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.777151 (rank : 34) | θ value | 2.97466e-19 (rank : 29) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.775505 (rank : 35) | θ value | 9.8567e-31 (rank : 10) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.774616 (rank : 36) | θ value | 1.13037e-18 (rank : 31) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.772566 (rank : 37) | θ value | 3.74554e-30 (rank : 11) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.760724 (rank : 38) | θ value | 3.89403e-11 (rank : 51) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.746846 (rank : 39) | θ value | 8.93572e-24 (rank : 16) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.731804 (rank : 40) | θ value | 4.02038e-16 (rank : 38) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.722098 (rank : 41) | θ value | 1.69304e-06 (rank : 59) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.708400 (rank : 42) | θ value | 4.30538e-10 (rank : 53) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.692885 (rank : 43) | θ value | 1.68911e-14 (rank : 44) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.692549 (rank : 44) | θ value | 2.20605e-14 (rank : 45) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.690530 (rank : 45) | θ value | 6.00763e-12 (rank : 48) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.688163 (rank : 46) | θ value | 1.33837e-11 (rank : 49) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.686750 (rank : 47) | θ value | 6.64225e-11 (rank : 52) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.685486 (rank : 48) | θ value | 1.74796e-11 (rank : 50) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.683411 (rank : 49) | θ value | 3.18553e-13 (rank : 46) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.682501 (rank : 50) | θ value | 1.58096e-12 (rank : 47) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.673915 (rank : 51) | θ value | 5.62301e-10 (rank : 54) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.665972 (rank : 52) | θ value | 8.11959e-09 (rank : 57) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.645902 (rank : 53) | θ value | 3.19293e-05 (rank : 61) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.645152 (rank : 54) | θ value | 1.63604e-09 (rank : 56) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.643215 (rank : 55) | θ value | 1.63604e-09 (rank : 55) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.638492 (rank : 56) | θ value | 1.29631e-06 (rank : 58) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP14_MOUSE
|
||||||
| NC score | 0.612226 (rank : 57) | θ value | 1.09739e-05 (rank : 60) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP28_HUMAN
|
||||||
| NC score | 0.589569 (rank : 58) | θ value | 2.60593e-15 (rank : 42) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.583392 (rank : 59) | θ value | 4.44505e-15 (rank : 43) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP25_MOUSE
|
||||||
| NC score | 0.583221 (rank : 60) | θ value | 7.32683e-18 (rank : 33) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP25_HUMAN
|
||||||
| NC score | 0.581865 (rank : 61) | θ value | 1.92812e-18 (rank : 32) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.567420 (rank : 62) | θ value | 0.00228821 (rank : 66) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.548285 (rank : 63) | θ value | 0.00298849 (rank : 67) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.517281 (rank : 64) | θ value | 0.00390308 (rank : 68) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.494594 (rank : 65) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.490359 (rank : 66) | θ value | 0.00035302 (rank : 63) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.482722 (rank : 67) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.480188 (rank : 68) | θ value | 5.44631e-05 (rank : 62) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.474780 (rank : 69) | θ value | 0.000602161 (rank : 64) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.474383 (rank : 70) | θ value | 0.000602161 (rank : 65) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP29_MOUSE
|
||||||
| NC score | 0.464360 (rank : 71) | θ value | 0.0193708 (rank : 69) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.445758 (rank : 72) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.373467 (rank : 73) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
BRAP_MOUSE
|
||||||
| NC score | 0.095952 (rank : 74) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
BRAP_HUMAN
|
||||||
| NC score | 0.084778 (rank : 75) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
UCRP_MOUSE
|
||||||
| NC score | 0.073435 (rank : 76) | θ value | 0.21417 (rank : 71) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64339, Q8K0H3 | Gene names | Isg15, G1p2, Ucrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin cross-reactive protein precursor (Interferon-stimulated protein 15) (IP17). | |||||
|
GGNB1_MOUSE
|
||||||
| NC score | 0.067661 (rank : 77) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6K1E7, Q8C5T9 | Gene names | Ggnbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
HDAC6_MOUSE
|
||||||
| NC score | 0.065237 (rank : 78) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
HDAC6_HUMAN
|
||||||
| NC score | 0.063102 (rank : 79) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
UCRP_HUMAN
|
||||||
| NC score | 0.055997 (rank : 80) | θ value | 1.06291 (rank : 80) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05161, Q7Z2G2, Q96GF0 | Gene names | ISG15, G1P2, UCRP | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced 17 kDa protein precursor [Contains: Ubiquitin cross-reactive protein (hUCRP) (Interferon-induced 15 kDa protein)]. | |||||
|
ZRAB2_HUMAN
|
||||||
| NC score | 0.054260 (rank : 81) | θ value | 0.813845 (rank : 76) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
GGNB1_HUMAN
|
||||||
| NC score | 0.052532 (rank : 82) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5YKI7, Q5YKI8 | Gene names | GGNBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
UBP54_MOUSE
|
||||||
| NC score | 0.052354 (rank : 83) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP54_HUMAN
|
||||||
| NC score | 0.052200 (rank : 84) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.036551 (rank : 85) | θ value | 0.21417 (rank : 70) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
SIAL_HUMAN
|
||||||
| NC score | 0.034268 (rank : 86) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |

|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
ARS2_HUMAN
|
||||||
| NC score | 0.033640 (rank : 87) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
ARS2_MOUSE
|
||||||
| NC score | 0.024429 (rank : 88) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.021413 (rank : 89) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.021281 (rank : 90) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
DGKH_HUMAN
|
||||||
| NC score | 0.019220 (rank : 91) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.018940 (rank : 92) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
TF3C5_HUMAN
|
||||||
| NC score | 0.018435 (rank : 93) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Q8, Q8N2U7, Q8N857, Q96GD9, Q9H4P2 | Gene names | GTF3C5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 5 (Transcription factor IIIC-epsilon subunit) (TF3C-epsilon) (TFIIIC 63 kDa subunit) (TFIIIC63). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.018369 (rank : 94) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.018304 (rank : 95) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.016962 (rank : 96) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.015791 (rank : 97) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.015773 (rank : 98) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
STAC3_MOUSE
|
||||||
| NC score | 0.015584 (rank : 99) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BZ71 | Gene names | Stac3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
NO40_MOUSE
|
||||||
| NC score | 0.015196 (rank : 100) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESX4 | Gene names | Zcchc17, Ps1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein). | |||||
|
TF3C5_MOUSE
|
||||||
| NC score | 0.014334 (rank : 101) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2T8, Q8BLE3, Q8R0D0 | Gene names | Gtf3c5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 5 (Transcription factor IIIC-epsilon subunit) (TF3C-epsilon) (TFIIIC 63 kDa subunit) (TFIIIC63). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.013489 (rank : 102) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.013428 (rank : 103) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.013220 (rank : 104) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.013203 (rank : 105) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.012997 (rank : 106) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
GDAP1_MOUSE
|
||||||
| NC score | 0.010677 (rank : 107) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88741, Q8C7Q5, Q9CTN2 | Gene names | Gdap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.010253 (rank : 108) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
ALBU_HUMAN
|
||||||
| NC score | 0.010165 (rank : 109) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02768, O95574, P04277, Q13140, Q68DN5, Q6UXK4, Q9P157, Q9P1I7, Q9UHS3, Q9UJZ0 | Gene names | ALB | |||
|
Domain Architecture |

|
|||||
| Description | Serum albumin precursor. | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.009141 (rank : 110) | θ value | 1.06291 (rank : 78) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.008751 (rank : 111) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
HDGF_MOUSE
|
||||||
| NC score | 0.008585 (rank : 112) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
PSD7_MOUSE
|
||||||
| NC score | 0.008263 (rank : 113) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26516, Q3TW61, Q8C0I8 | Gene names | Psmd7, Mov-34, Mov34 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein). | |||||
|
CAN3_HUMAN
|
||||||
| NC score | 0.008229 (rank : 114) | θ value | 0.62314 (rank : 74) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.007847 (rank : 115) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
AKP8L_HUMAN
|
||||||
| NC score | 0.007329 (rank : 116) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULX6, O94792, Q96J58, Q9NRQ0, Q9UGM0 | Gene names | AKAP8L, NAKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8-like (AKAP8-like protein) (Neighbor of A- kinase-anchoring protein 95) (Neighbor of AKAP95) (Homologous to AKAP95 protein) (HA95) (Helicase A-binding protein 95) (HAP95). | |||||
|
RNF8_HUMAN
|
||||||
| NC score | 0.007006 (rank : 117) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76064 | Gene names | RNF8, KIAA0646 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8). | |||||
|
CSTN1_HUMAN
|
||||||
| NC score | 0.006870 (rank : 118) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94985 | Gene names | CLSTN1, CS1, KIAA0911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.006166 (rank : 119) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.005388 (rank : 120) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.005375 (rank : 121) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
FBXW7_HUMAN
|
||||||
| NC score | 0.004378 (rank : 122) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.004351 (rank : 123) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.004222 (rank : 124) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.003213 (rank : 125) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.000281 (rank : 126) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
BRWD1_HUMAN
|
||||||
| NC score | -0.000067 (rank : 127) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | -0.000704 (rank : 128) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | -0.000946 (rank : 129) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MINK1_MOUSE
|
||||||
| NC score | -0.005861 (rank : 130) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MKNK2_MOUSE
|
||||||
| NC score | -0.005909 (rank : 131) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDB0, O08606, Q75PY0, Q9D893 | Gene names | Mknk2, Mnk2 | |||
|
Domain Architecture |

|
|||||
| Description | MAP kinase-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (MAP kinase signal-integrating kinase 2) (Mnk2). | |||||
|
KPCT_MOUSE
|
||||||
| NC score | -0.006034 (rank : 132) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 124 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02111 | Gene names | Prkcq, Pkcq | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||