Please be patient as the page loads
|
UBP28_MOUSE
|
||||||
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBP25_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.966771 (rank : 4) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP25_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.969245 (rank : 3) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP28_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.986107 (rank : 2) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 178 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 5) | NC score | 0.586723 (rank : 5) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 6) | NC score | 0.583392 (rank : 6) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 7) | NC score | 0.509641 (rank : 9) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 8) | NC score | 0.507887 (rank : 11) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 9) | NC score | 0.483311 (rank : 21) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 10) | NC score | 0.496272 (rank : 15) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 11) | NC score | 0.516344 (rank : 8) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 12) | NC score | 0.480352 (rank : 22) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 13) | NC score | 0.495522 (rank : 16) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 14) | NC score | 0.472004 (rank : 24) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 15) | NC score | 0.504779 (rank : 13) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 16) | NC score | 0.509158 (rank : 10) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 17) | NC score | 0.536305 (rank : 7) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 18) | NC score | 0.505165 (rank : 12) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 19) | NC score | 0.442540 (rank : 25) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 20) | NC score | 0.504077 (rank : 14) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 21) | NC score | 0.343076 (rank : 59) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 22) | NC score | 0.431480 (rank : 26) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 23) | NC score | 0.427765 (rank : 28) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 24) | NC score | 0.491842 (rank : 19) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 25) | NC score | 0.491842 (rank : 20) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 26) | NC score | 0.380378 (rank : 49) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 27) | NC score | 0.494840 (rank : 17) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 28) | NC score | 0.493624 (rank : 18) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 29) | NC score | 0.474823 (rank : 23) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 30) | NC score | 0.343329 (rank : 58) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 31) | NC score | 0.360797 (rank : 53) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 32) | NC score | 0.428416 (rank : 27) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 33) | NC score | 0.422964 (rank : 30) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 34) | NC score | 0.405310 (rank : 33) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP29_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 35) | NC score | 0.371543 (rank : 52) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 36) | NC score | 0.397971 (rank : 42) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 37) | NC score | 0.379736 (rank : 51) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 38) | NC score | 0.410767 (rank : 31) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 39) | NC score | 0.312421 (rank : 66) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 40) | NC score | 0.307162 (rank : 68) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 41) | NC score | 0.294484 (rank : 69) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 42) | NC score | 0.354022 (rank : 56) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 43) | NC score | 0.357197 (rank : 55) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 44) | NC score | 0.404633 (rank : 35) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP14_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 45) | NC score | 0.392200 (rank : 43) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 46) | NC score | 0.398877 (rank : 41) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 47) | NC score | 0.342465 (rank : 60) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 48) | NC score | 0.400809 (rank : 40) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 49) | NC score | 0.308173 (rank : 67) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 50) | NC score | 0.404477 (rank : 36) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 51) | NC score | 0.381154 (rank : 46) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 52) | NC score | 0.424653 (rank : 29) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 53) | NC score | 0.404704 (rank : 34) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 54) | NC score | 0.400927 (rank : 39) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 55) | NC score | 0.409971 (rank : 32) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
TLE2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 56) | NC score | 0.017381 (rank : 110) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
UBP20_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 57) | NC score | 0.401037 (rank : 38) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 58) | NC score | 0.402154 (rank : 37) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 59) | NC score | 0.332147 (rank : 64) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.339327 (rank : 61) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 61) | NC score | 0.380947 (rank : 48) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 62) | NC score | 0.025101 (rank : 88) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 63) | NC score | 0.381051 (rank : 47) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 64) | NC score | 0.379954 (rank : 50) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 65) | NC score | 0.040496 (rank : 75) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 0.125558 (rank : 66) | NC score | 0.391768 (rank : 44) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
CEP68_HUMAN
|
||||||
| θ value | 0.163984 (rank : 67) | NC score | 0.040779 (rank : 74) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
ZCH14_MOUSE
|
||||||
| θ value | 0.21417 (rank : 68) | NC score | 0.035977 (rank : 79) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 69) | NC score | 0.002067 (rank : 171) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MYC_MOUSE
|
||||||
| θ value | 0.279714 (rank : 70) | NC score | 0.033233 (rank : 81) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
REV1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 71) | NC score | 0.029931 (rank : 83) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 0.279714 (rank : 72) | NC score | 0.333434 (rank : 63) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 0.279714 (rank : 73) | NC score | 0.333726 (rank : 62) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 74) | NC score | 0.013076 (rank : 125) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
UB17L_HUMAN
|
||||||
| θ value | 0.365318 (rank : 75) | NC score | 0.390936 (rank : 45) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 0.47712 (rank : 76) | NC score | 0.025234 (rank : 87) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.023136 (rank : 92) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
HIRP5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 78) | NC score | 0.037077 (rank : 77) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZ23, Q9D1B7 | Gene names | Hirip5 | |||
|
Domain Architecture |
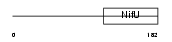
|
|||||
| Description | HIRA-interacting protein 5 (mHIRIP5). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 79) | NC score | 0.357981 (rank : 54) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.022398 (rank : 94) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.029591 (rank : 84) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 82) | NC score | 0.017271 (rank : 111) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 83) | NC score | 0.019592 (rank : 101) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PRRT1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 84) | NC score | 0.039108 (rank : 76) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 85) | NC score | 0.240166 (rank : 73) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 86) | NC score | 0.245941 (rank : 72) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
CM018_HUMAN
|
||||||
| θ value | 1.06291 (rank : 87) | NC score | 0.029405 (rank : 85) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H714, Q5W051, Q6PJ74, Q6PK94, Q86XH7, Q8N5J6 | Gene names | C13orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf18. | |||||
|
CMTA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.019380 (rank : 103) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 89) | NC score | 0.015348 (rank : 117) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 90) | NC score | 0.034339 (rank : 80) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
STAT6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 91) | NC score | 0.014723 (rank : 118) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
DOCK6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.013746 (rank : 123) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
MYC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.028137 (rank : 86) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
PCY1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 94) | NC score | 0.018356 (rank : 107) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49586, Q542W4 | Gene names | Pcyt1a, Ctpct, Pcyt1 | |||
|
Domain Architecture |
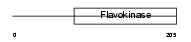
|
|||||
| Description | Choline-phosphate cytidylyltransferase A (EC 2.7.7.15) (Phosphorylcholine transferase A) (CTP:phosphocholine cytidylyltransferase A) (CT A) (CCT A) (CCT-alpha). | |||||
|
STAR3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 95) | NC score | 0.024590 (rank : 89) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14849, Q53Y53, Q96HM9 | Gene names | STARD3, CAB1, MLN64 | |||
|
Domain Architecture |
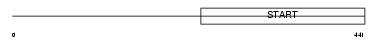
|
|||||
| Description | StAR-related lipid transfer protein 3 (StARD3) (START domain- containing protein 3) (Metastatic lymph node protein 64) (Protein MLN 64) (Protein CAB1). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | 0.006398 (rank : 155) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
PRDM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | -0.001672 (rank : 178) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
PXK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.018098 (rank : 109) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7A4, Q3BCH4, Q3BCH5, Q3BCH6, Q3BDW1, Q45L83, Q59EX3, Q6PK17, Q6ZN39, Q96CA3, Q96R07, Q9NXB8 | Gene names | PXK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
ATF4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.021096 (rank : 99) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
CD68_MOUSE
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.021978 (rank : 95) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31996 | Gene names | Cd68 | |||
|
Domain Architecture |
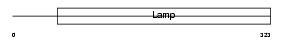
|
|||||
| Description | Macrosialin precursor (CD68 antigen). | |||||
|
CEP55_HUMAN
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.019497 (rank : 102) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.014181 (rank : 122) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
RUSC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.015476 (rank : 115) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.007220 (rank : 147) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
STAR3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.023016 (rank : 93) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61542 | Gene names | Stard3, Es64, Mln64 | |||
|
Domain Architecture |
|
|||||
| Description | StAR-related lipid transfer protein 3 (StARD3) (START domain- containing protein 3) (Protein MLN 64) (Protein ES 64). | |||||
|
TLR6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.008812 (rank : 141) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPW9, Q9WTQ4 | Gene names | Tlr6 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 6 precursor. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | 0.005514 (rank : 159) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 108) | NC score | 0.018790 (rank : 105) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.016258 (rank : 114) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
BNC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.016437 (rank : 113) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.014680 (rank : 119) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CLAP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.014431 (rank : 121) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CT096_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.021265 (rank : 97) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NUD7, Q8N840, Q8NAX5 | Gene names | C20orf96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf96. | |||||
|
DGKD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.009837 (rank : 138) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
DHB5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.007119 (rank : 150) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70694 | Gene names | Akr1c6, Hsd17b5 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17 beta-dehydrogenase 5 (EC 1.1.1.-) (17-beta-HSD 5). | |||||
|
DSC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.002133 (rank : 170) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02487 | Gene names | DSC2, DSC3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Desmosomal glycoprotein II and III) (Desmocollin-3). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.010640 (rank : 135) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
K0586_HUMAN
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.021560 (rank : 96) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | 0.000546 (rank : 174) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
NOL6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 120) | NC score | 0.036052 (rank : 78) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5K4, Q6PAN8, Q8C6V4, Q8R5K3, Q8VCG0, Q8WTY7 | Gene names | Nol6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
PDLI5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 121) | NC score | 0.007675 (rank : 146) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 122) | NC score | 0.010610 (rank : 136) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRRT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 123) | NC score | 0.032538 (rank : 82) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
TBCE_MOUSE
|
||||||
| θ value | 3.0926 (rank : 124) | NC score | 0.021230 (rank : 98) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CIV8 | Gene names | Tbce | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin-specific chaperone E (Tubulin-folding cofactor E). | |||||
|
TLR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 125) | NC score | 0.008769 (rank : 142) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPQ1, Q9EPW5 | Gene names | Tlr1 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 1 precursor (Toll/interleukin-1 receptor-like protein) (TIL) (CD281 antigen). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 126) | NC score | 0.292529 (rank : 70) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 127) | NC score | -0.000540 (rank : 175) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
BNC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.015457 (rank : 116) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.010717 (rank : 134) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.011771 (rank : 131) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.016692 (rank : 112) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.019633 (rank : 100) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TRAF7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.004887 (rank : 163) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6Q0C0, Q9H073 | Gene names | TRAF7, RFWD1, RNF119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.-) (TNF receptor- associated factor 7) (RING finger and WD repeat domain protein 1) (RING finger protein 119). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 134) | NC score | 0.288885 (rank : 71) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 135) | NC score | -0.001436 (rank : 177) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.014451 (rank : 120) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.005949 (rank : 156) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
DRP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.008142 (rank : 145) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.006439 (rank : 154) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
LBXCO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.010428 (rank : 137) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BX46, Q3UYA4, Q5W8I2, Q8C0T2 | Gene names | Lbxcor1, Corl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1 (Transcriptional corepressor Corl1). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.018550 (rank : 106) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
RABE2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.009692 (rank : 139) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
SPRE3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.011574 (rank : 132) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.018309 (rank : 108) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.019173 (rank : 104) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.004813 (rank : 164) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.008208 (rank : 144) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AT2B3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.011142 (rank : 133) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.002648 (rank : 167) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.012745 (rank : 128) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GPSM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.007124 (rank : 149) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.013071 (rank : 126) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.004205 (rank : 165) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.005781 (rank : 157) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.001901 (rank : 172) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.000569 (rank : 173) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
UBL7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.024226 (rank : 90) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96S82, Q96I03 | Gene names | UBL7, BMSCUBP | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-like protein 7 (Ubiquitin-like protein SB132) (Bone marrow stromal cell ubiquitin-like protein) (BMSC-UbP). | |||||
|
UBL7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.024138 (rank : 91) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.315854 (rank : 65) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
ULK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | -0.000999 (rank : 176) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
ARHGG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.003183 (rank : 166) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.005581 (rank : 158) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.005405 (rank : 161) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CASR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.002419 (rank : 169) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
DMWD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.006463 (rank : 153) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08274 | Gene names | Dmwd, Dm9 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophia myotonica WD repeat-containing protein (Dystrophia myotonica-containing WD repeat motif protein) (DMR-N9 protein). | |||||
|
EAF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.012689 (rank : 129) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
EAF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.012920 (rank : 127) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D4C5 | Gene names | Eaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
GRP78_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.002571 (rank : 168) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
HDA11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.007025 (rank : 151) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WA3 | Gene names | Hdac11 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 11 (HD11). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.006613 (rank : 152) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.008560 (rank : 143) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
K0391_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.013317 (rank : 124) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15091 | Gene names | KIAA0391 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0391. | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.012291 (rank : 130) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.009493 (rank : 140) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PCGF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.005461 (rank : 160) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
PDLI5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.005182 (rank : 162) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CI51, Q9QYN0, Q9QYN1, Q9QYN2 | Gene names | Pdlim5, Enh | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
TEF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.007163 (rank : 148) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | -0.002171 (rank : 179) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.343846 (rank : 57) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 178 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_HUMAN
|
||||||
| NC score | 0.986107 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP25_MOUSE
|
||||||
| NC score | 0.969245 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP25_HUMAN
|
||||||
| NC score | 0.966771 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.586723 (rank : 5) | θ value | 8.95645e-16 (rank : 5) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.583392 (rank : 6) | θ value | 4.44505e-15 (rank : 6) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.536305 (rank : 7) | θ value | 2.61198e-07 (rank : 17) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.516344 (rank : 8) | θ value | 3.64472e-09 (rank : 11) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.509641 (rank : 9) | θ value | 1.33837e-11 (rank : 7) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.509158 (rank : 10) | θ value | 6.87365e-08 (rank : 16) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.507887 (rank : 11) | θ value | 6.64225e-11 (rank : 8) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.505165 (rank : 12) | θ value | 3.41135e-07 (rank : 18) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.504779 (rank : 13) | θ value | 5.26297e-08 (rank : 15) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.504077 (rank : 14) | θ value | 4.45536e-07 (rank : 20) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.496272 (rank : 15) | θ value | 2.13673e-09 (rank : 10) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.495522 (rank : 16) | θ value | 4.76016e-09 (rank : 13) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.494840 (rank : 17) | θ value | 3.19293e-05 (rank : 27) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.493624 (rank : 18) | θ value | 4.1701e-05 (rank : 28) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.491842 (rank : 19) | θ value | 1.87187e-05 (rank : 24) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.491842 (rank : 20) | θ value | 1.87187e-05 (rank : 25) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.483311 (rank : 21) | θ value | 1.25267e-09 (rank : 9) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.480352 (rank : 22) | θ value | 3.64472e-09 (rank : 12) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.474823 (rank : 23) | θ value | 0.00020696 (rank : 29) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.472004 (rank : 24) | θ value | 1.06045e-08 (rank : 14) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.442540 (rank : 25) | θ value | 4.45536e-07 (rank : 19) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.431480 (rank : 26) | θ value | 1.43324e-05 (rank : 22) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.428416 (rank : 27) | θ value | 0.000270298 (rank : 32) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.427765 (rank : 28) | θ value | 1.87187e-05 (rank : 23) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.424653 (rank : 29) | θ value | 0.00869519 (rank : 52) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.422964 (rank : 30) | θ value | 0.000602161 (rank : 33) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.410767 (rank : 31) | θ value | 0.00175202 (rank : 38) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.409971 (rank : 32) | θ value | 0.0193708 (rank : 55) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.405310 (rank : 33) | θ value | 0.000786445 (rank : 34) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.404704 (rank : 34) | θ value | 0.0113563 (rank : 53) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.404633 (rank : 35) | θ value | 0.00390308 (rank : 44) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.404477 (rank : 36) | θ value | 0.00869519 (rank : 50) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.402154 (rank : 37) | θ value | 0.0431538 (rank : 58) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.401037 (rank : 38) | θ value | 0.0431538 (rank : 57) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.400927 (rank : 39) | θ value | 0.0148317 (rank : 54) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.400809 (rank : 40) | θ value | 0.00509761 (rank : 48) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.398877 (rank : 41) | θ value | 0.00390308 (rank : 46) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.397971 (rank : 42) | θ value | 0.00102713 (rank : 36) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP14_MOUSE
|
||||||
| NC score | 0.392200 (rank : 43) | θ value | 0.00390308 (rank : 45) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.391768 (rank : 44) | θ value | 0.125558 (rank : 66) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.390936 (rank : 45) | θ value | 0.365318 (rank : 75) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.381154 (rank : 46) | θ value | 0.00869519 (rank : 51) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.381051 (rank : 47) | θ value | 0.0961366 (rank : 63) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.380947 (rank : 48) | θ value | 0.0736092 (rank : 61) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.380378 (rank : 49) | θ value | 2.44474e-05 (rank : 26) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.379954 (rank : 50) | θ value | 0.0961366 (rank : 64) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.379736 (rank : 51) | θ value | 0.00134147 (rank : 37) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP29_MOUSE
|
||||||
| NC score | 0.371543 (rank : 52) | θ value | 0.00102713 (rank : 35) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.360797 (rank : 53) | θ value | 0.000270298 (rank : 31) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.357981 (rank : 54) | θ value | 0.62314 (rank : 79) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.357197 (rank : 55) | θ value | 0.00298849 (rank : 43) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.354022 (rank : 56) | θ value | 0.00298849 (rank : 42) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.343846 (rank : 57) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.343329 (rank : 58) | θ value | 0.000270298 (rank : 30) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.343076 (rank : 59) | θ value | 6.43352e-06 (rank : 21) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.342465 (rank : 60) | θ value | 0.00509761 (rank : 47) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.339327 (rank : 61) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.333726 (rank : 62) | θ value | 0.279714 (rank : 73) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.333434 (rank : 63) | θ value | 0.279714 (rank : 72) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.332147 (rank : 64) | θ value | 0.0563607 (rank : 59) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.315854 (rank : 65) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.312421 (rank : 66) | θ value | 0.00175202 (rank : 39) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.308173 (rank : 67) | θ value | 0.00665767 (rank : 49) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.307162 (rank : 68) | θ value | 0.00228821 (rank : 40) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.294484 (rank : 69) | θ value | 0.00228821 (rank : 41) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.292529 (rank : 70) | θ value | 3.0926 (rank : 126) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.288885 (rank : 71) | θ value | 4.03905 (rank : 134) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.245941 (rank : 72) | θ value | 0.813845 (rank : 86) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.240166 (rank : 73) | θ value | 0.813845 (rank : 85) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
CEP68_HUMAN
|
||||||
| NC score | 0.040779 (rank : 74) | θ value | 0.163984 (rank : 67) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.040496 (rank : 75) | θ value | 0.125558 (rank : 65) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
PRRT1_MOUSE
|
||||||
| NC score | 0.039108 (rank : 76) | θ value | 0.813845 (rank : 84) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35449 | Gene names | Prrt1, Ng5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
HIRP5_MOUSE
|
||||||
| NC score | 0.037077 (rank : 77) | θ value | 0.62314 (rank : 78) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZ23, Q9D1B7 | Gene names | Hirip5 | |||
|
Domain Architecture |

|
|||||
| Description | HIRA-interacting protein 5 (mHIRIP5). | |||||
|
NOL6_MOUSE
|
||||||
| NC score | 0.036052 (rank : 78) | θ value | 3.0926 (rank : 120) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5K4, Q6PAN8, Q8C6V4, Q8R5K3, Q8VCG0, Q8WTY7 | Gene names | Nol6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
ZCH14_MOUSE
|
||||||
| NC score | 0.035977 (rank : 79) | θ value | 0.21417 (rank : 68) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.034339 (rank : 80) | θ value | 1.06291 (rank : 90) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.033233 (rank : 81) | θ value | 0.279714 (rank : 70) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
PRRT1_HUMAN
|
||||||
| NC score | 0.032538 (rank : 82) | θ value | 3.0926 (rank : 123) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
REV1_HUMAN
|
||||||
| NC score | 0.029931 (rank : 83) | θ value | 0.279714 (rank : 71) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.029591 (rank : 84) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
CM018_HUMAN
|
||||||
| NC score | 0.029405 (rank : 85) | θ value | 1.06291 (rank : 87) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H714, Q5W051, Q6PJ74, Q6PK94, Q86XH7, Q8N5J6 | Gene names | C13orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf18. | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.028137 (rank : 86) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.025234 (rank : 87) | θ value | 0.47712 (rank : 76) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.025101 (rank : 88) | θ value | 0.0961366 (rank : 62) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
STAR3_HUMAN
|
||||||
| NC score | 0.024590 (rank : 89) | θ value | 1.38821 (rank : 95) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14849, Q53Y53, Q96HM9 | Gene names | STARD3, CAB1, MLN64 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 3 (StARD3) (START domain- containing protein 3) (Metastatic lymph node protein 64) (Protein MLN 64) (Protein CAB1). | |||||
|
UBL7_HUMAN
|
||||||
| NC score | 0.024226 (rank : 90) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96S82, Q96I03 | Gene names | UBL7, BMSCUBP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 7 (Ubiquitin-like protein SB132) (Bone marrow stromal cell ubiquitin-like protein) (BMSC-UbP). | |||||
|
UBL7_MOUSE
|
||||||
| NC score | 0.024138 (rank : 91) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W67, Q9D7P5 | Gene names | Ubl7 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like protein 7. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.023136 (rank : 92) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
STAR3_MOUSE
|
||||||
| NC score | 0.023016 (rank : 93) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61542 | Gene names | Stard3, Es64, Mln64 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 3 (StARD3) (START domain- containing protein 3) (Protein MLN 64) (Protein ES 64). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.022398 (rank : 94) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CD68_MOUSE
|
||||||
| NC score | 0.021978 (rank : 95) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31996 | Gene names | Cd68 | |||
|
Domain Architecture |

|
|||||
| Description | Macrosialin precursor (CD68 antigen). | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.021560 (rank : 96) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
CT096_HUMAN
|
||||||
| NC score | 0.021265 (rank : 97) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NUD7, Q8N840, Q8NAX5 | Gene names | C20orf96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf96. | |||||
|
TBCE_MOUSE
|
||||||
| NC score | 0.021230 (rank : 98) | θ value | 3.0926 (rank : 124) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CIV8 | Gene names | Tbce | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin-specific chaperone E (Tubulin-folding cofactor E). | |||||
|
ATF4_MOUSE
|
||||||
| NC score | 0.021096 (rank : 99) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.019633 (rank : 100) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.019592 (rank : 101) | θ value | 0.813845 (rank : 83) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
CEP55_HUMAN
|
||||||
| NC score | 0.019497 (rank : 102) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q53EZ4, Q32WF5, Q3MV20, Q5VY28, Q6N034, Q96H32, Q9NVS7 | Gene names | CEP55, C10orf3, URCC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55 (Up-regulated in colon cancer 6). | |||||
|
CMTA2_HUMAN
|
||||||
| NC score | 0.019380 (rank : 103) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.019173 (rank : 104) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.018790 (rank : 105) | θ value | 2.36792 (rank : 108) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.018550 (rank : 106) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
PCY1A_MOUSE
|
||||||
| NC score | 0.018356 (rank : 107) | θ value | 1.38821 (rank : 94) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49586, Q542W4 | Gene names | Pcyt1a, Ctpct, Pcyt1 | |||
|
Domain Architecture |

|
|||||
| Description | Choline-phosphate cytidylyltransferase A (EC 2.7.7.15) (Phosphorylcholine transferase A) (CTP:phosphocholine cytidylyltransferase A) (CT A) (CCT A) (CCT-alpha). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.018309 (rank : 108) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
PXK_HUMAN
|
||||||
| NC score | 0.018098 (rank : 109) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z7A4, Q3BCH4, Q3BCH5, Q3BCH6, Q3BDW1, Q45L83, Q59EX3, Q6PK17, Q6ZN39, Q96CA3, Q96R07, Q9NXB8 | Gene names | PXK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
TLE2_HUMAN
|
||||||
| NC score | 0.017381 (rank : 110) | θ value | 0.0431538 (rank : 56) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.017271 (rank : 111) | θ value | 0.813845 (rank : 82) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.016692 (rank : 112) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BNC2_HUMAN
|
||||||
| NC score | 0.016437 (rank : 113) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.016258 (rank : 114) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
RUSC2_HUMAN
|
||||||
| NC score | 0.015476 (rank : 115) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N2Y8, O15080, Q6P1W7 | Gene names | RUSC2, KIAA0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
BNC2_MOUSE
|
||||||
| NC score | 0.015457 (rank : 116) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.015348 (rank : 117) | θ value | 1.06291 (rank : 89) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
STAT6_HUMAN
|
||||||
| NC score | 0.014723 (rank : 118) | θ value | 1.06291 (rank : 91) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.014680 (rank : 119) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.014451 (rank : 120) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CLAP2_MOUSE
|
||||||
| NC score | 0.014431 (rank : 121) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.014181 (rank : 122) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
DOCK6_HUMAN
|
||||||
| NC score | 0.013746 (rank : 123) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
K0391_HUMAN
|
||||||
| NC score | 0.013317 (rank : 124) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15091 | Gene names | KIAA0391 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0391. | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.013076 (rank : 125) | θ value | 0.279714 (rank : 74) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.013071 (rank : 126) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
EAF1_MOUSE
|
||||||
| NC score | 0.012920 (rank : 127) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D4C5 | Gene names | Eaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.012745 (rank : 128) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
EAF1_HUMAN
|
||||||
| NC score | 0.012689 (rank : 129) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.012291 (rank : 130) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.011771 (rank : 131) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SPRE3_MOUSE
|
||||||
| NC score | 0.011574 (rank : 132) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
AT2B3_HUMAN
|
||||||
| NC score | 0.011142 (rank : 133) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16720, Q12995, Q16858 | Gene names | ATP2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 3 (EC 3.6.3.8) (PMCA3) (Plasma membrane calcium pump isoform 3) (Plasma membrane calcium ATPase isoform 3). | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.010717 (rank : 134) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.010640 (rank : 135) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.010610 (rank : 136) | θ value | 3.0926 (rank : 122) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
LBXCO_MOUSE
|
||||||
| NC score | 0.010428 (rank : 137) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BX46, Q3UYA4, Q5W8I2, Q8C0T2 | Gene names | Lbxcor1, Corl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1 (Transcriptional corepressor Corl1). | |||||
|
DGKD_HUMAN
|
||||||
| NC score | 0.009837 (rank : 138) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
RABE2_MOUSE
|
||||||
| NC score | 0.009692 (rank : 139) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.009493 (rank : 140) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
TLR6_MOUSE
|
||||||
| NC score | 0.008812 (rank : 141) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPW9, Q9WTQ4 | Gene names | Tlr6 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 6 precursor. | |||||
|
TLR1_MOUSE
|
||||||
| NC score | 0.008769 (rank : 142) | θ value | 3.0926 (rank : 125) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EPQ1, Q9EPW5 | Gene names | Tlr1 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 1 precursor (Toll/interleukin-1 receptor-like protein) (TIL) (CD281 antigen). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.008560 (rank : 143) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.008208 (rank : 144) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
DRP2_HUMAN
|
||||||
| NC score | 0.008142 (rank : 145) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
PDLI5_HUMAN
|
||||||
| NC score | 0.007675 (rank : 146) | θ value | 3.0926 (rank : 121) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.007220 (rank : 147) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
TEF_HUMAN
|
||||||
| NC score | 0.007163 (rank : 148) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
GPSM1_MOUSE
|
||||||
| NC score | 0.007124 (rank : 149) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6IR34, Q61366, Q8BUK4, Q8BX78, Q8R0R9 | Gene names | Gpsm1, Ags3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 1 (Activator of G-protein signaling 3). | |||||
|
DHB5_MOUSE
|
||||||
| NC score | 0.007119 (rank : 150) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70694 | Gene names | Akr1c6, Hsd17b5 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17 beta-dehydrogenase 5 (EC 1.1.1.-) (17-beta-HSD 5). | |||||
|
HDA11_MOUSE
|
||||||
| NC score | 0.007025 (rank : 151) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WA3 | Gene names | Hdac11 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 11 (HD11). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.006613 (rank : 152) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
DMWD_MOUSE
|
||||||
| NC score | 0.006463 (rank : 153) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08274 | Gene names | Dmwd, Dm9 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophia myotonica WD repeat-containing protein (Dystrophia myotonica-containing WD repeat motif protein) (DMR-N9 protein). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.006439 (rank : 154) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.006398 (rank : 155) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.005949 (rank : 156) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.005781 (rank : 157) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.005581 (rank : 158) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.005514 (rank : 159) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
PCGF2_MOUSE
|
||||||
| NC score | 0.005461 (rank : 160) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.005405 (rank : 161) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
PDLI5_MOUSE
|
||||||
| NC score | 0.005182 (rank : 162) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CI51, Q9QYN0, Q9QYN1, Q9QYN2 | Gene names | Pdlim5, Enh | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
TRAF7_HUMAN
|
||||||
| NC score | 0.004887 (rank : 163) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6Q0C0, Q9H073 | Gene names | TRAF7, RFWD1, RNF119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.-) (TNF receptor- associated factor 7) (RING finger and WD repeat domain protein 1) (RING finger protein 119). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.004813 (rank : 164) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.004205 (rank : 165) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
ARHGG_MOUSE
|
||||||
| NC score | 0.003183 (rank : 166) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.002648 (rank : 167) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
GRP78_HUMAN
|
||||||
| NC score | 0.002571 (rank : 168) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
CASR_HUMAN
|
||||||
| NC score | 0.002419 (rank : 169) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
DSC2_HUMAN
|
||||||
| NC score | 0.002133 (rank : 170) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02487 | Gene names | DSC2, DSC3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Desmosomal glycoprotein II and III) (Desmocollin-3). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.002067 (rank : 171) | θ value | 0.279714 (rank : 69) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.001901 (rank : 172) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.000569 (rank : 173) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.000546 (rank : 174) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
ULK2_HUMAN
|
||||||
| NC score | -0.000540 (rank : 175) | θ value | 3.0926 (rank : 127) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
ULK2_MOUSE
|
||||||
| NC score | -0.000999 (rank : 176) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | -0.001436 (rank : 177) | θ value | 4.03905 (rank : 135) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
PRDM1_MOUSE
|
||||||
| NC score | -0.001672 (rank : 178) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60636 | Gene names | Prdm1, Blimp1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | -0.002171 (rank : 179) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 178 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||