Please be patient as the page loads
|
CLAP2_MOUSE
|
||||||
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CLAP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.957978 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
CLAP2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991846 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O75122, Q7L8F6, Q8N6R6, Q9BQT3, Q9BQT4, Q9H7A3, Q9NSZ2 | Gene names | CLASP2, KIAA0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CLAP2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CAND1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 4) | NC score | 0.117960 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86VP6, O94918, Q6PIY4, Q8NDJ4, Q96JZ9, Q96T19, Q9BTC4, Q9H0G2, Q9P0H7, Q9UF85 | Gene names | CAND1, KIAA0829, TIP120, TIP120A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1) (TBP-interacting protein TIP120A) (TBP-interacting protein of 120 kDa A). | |||||
|
CKAP5_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 5) | NC score | 0.179794 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14008, Q14668 | Gene names | CKAP5, KIAA0097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 5 (Colonic and hepatic tumor over- expressed protein) (Ch-TOG protein). | |||||
|
CAND1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.115976 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQ38, Q6PFR0, Q9CV45 | Gene names | Cand1, D10Ertd516e, Kiaa0829 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1). | |||||
|
IPO4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.117744 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TEX9, Q86TZ9, Q8NCG8, Q96SJ3, Q9BTI4, Q9H5L0 | Gene names | IPO4, IMP4B, RANBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-4 (Importin 4b) (Imp4b) (Ran-binding protein 4) (RanBP4). | |||||
|
FOXN3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.048444 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q499D0, Q8C5N8, Q9CU83 | Gene names | Ches1, Foxn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.048173 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
FOXN3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.045879 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00409, Q96II7, Q9UIE7 | Gene names | CHES1, FOXN3 | |||
|
Domain Architecture |
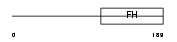
|
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.045752 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
UB2J1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.052063 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJZ4, Q9DC92 | Gene names | Ube2j1, Ncube1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1). | |||||
|
CAND2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.089139 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75155 | Gene names | CAND2, KIAA0667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2). | |||||
|
EPIPL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.039257 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0W0 | Gene names | Eppk1 | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin. | |||||
|
TEX10_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.072296 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXF1, Q5T722, Q5T723, Q8NCN8, Q8TDY0 | Gene names | TEX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 10 protein. | |||||
|
IPO4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.096806 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.018995 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
NR1H4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.020337 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RI1, Q8NFP5, Q8NFP6, Q92943 | Gene names | NR1H4, BAR, FXR, HRR1, RIP14 | |||
|
Domain Architecture |

|
|||||
| Description | Bile acid receptor (Farnesoid X-activated receptor) (Farnesol receptor HRR-1) (Retinoid X receptor-interacting protein 14) (RXR-interacting protein 14). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.042925 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
CASR_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.015063 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
TSH3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.016589 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
ATG9A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.030028 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3C6, Q6P0N7, Q7Z317, Q7Z320, Q8NDK6, Q8WU65, Q9BVL5, Q9H6L1, Q9HAG7 | Gene names | ATG9A, APG9L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
MAF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.027429 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.011252 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
PRC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.041832 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.043513 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
K1949_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.028023 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NYC8, Q68CK8, Q6ZUJ6, Q8NDQ4, Q8TF52, Q9BRL9 | Gene names | KIAA1949 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949. | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.018223 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
OST6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.012276 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QI5, Q96RX7 | Gene names | HS3ST6, HS3ST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (h3-OST-6). | |||||
|
RBP26_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.017589 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
ZSWM5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.019456 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
CBP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.014780 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CD2L6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.004086 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BWD8, Q80TM1 | Gene names | Cdc2l6, Kiaa1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6). | |||||
|
K2027_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.024160 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
MAF_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.024685 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MIA40_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.034217 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEA4 | Gene names | Chchd4, Mia40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial intermembrane space import and assembly protein 40 (Coiled-coil-helix-coiled-coil-helix domain-containing protein 4). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.017218 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
ZRAB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.099008 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
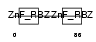
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
ANFC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.037067 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23582 | Gene names | NPPC, CNP2 | |||
|
Domain Architecture |
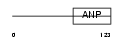
|
|||||
| Description | C-type natriuretic peptide precursor [Contains: CNP-22; CNP-29; CNP- 53]. | |||||
|
APC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.029610 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
BTBD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.009740 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX70, O60418, O75248, Q7Z5W0, Q96SX8, Q9NPS1, Q9NX81 | Gene names | BTBD2 | |||
|
Domain Architecture |
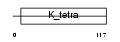
|
|||||
| Description | BTB/POZ domain-containing protein 2. | |||||
|
CDR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.022555 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97817 | Gene names | Cdr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2. | |||||
|
COVA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.027789 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
E41LB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.009701 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.022773 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MARK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.002567 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
SP8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.003602 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMJ8, Q8BWA3 | Gene names | Sp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp8. | |||||
|
TIRAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.042069 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58753, Q8N5E5 | Gene names | TIRAP, MAL | |||
|
Domain Architecture |
|
|||||
| Description | Toll-interleukin 1 receptor domain-containing adapter protein (TIR domain-containing adapter protein) (MyD88 adapter-like protein) (Adaptor protein Wyatt). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.014431 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
FIZ1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.001750 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTJ4, Q91W14, Q9CTG3 | Gene names | Fiz1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flt3-interacting zinc finger protein 1. | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.012839 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
TLN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.011279 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
YYAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.015983 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H869, Q5VYZ4, Q5VYZ7, Q7L4C3, Q7L5E2, Q8IXA6, Q8TEW5, Q8TF04, Q96HB6, Q9BQ64, Q9NV84 | Gene names | YY1AP1, HCCA2, YY1AP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YY1-associated protein 1 (Hepatocellular carcinoma susceptibility protein) (Hepatocellular carcinoma-associated protein 2). | |||||
|
CF152_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.009672 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
GCP60_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.013093 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
GP125_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.013261 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWK6, Q6UXK9, Q86SQ5, Q8TC55 | Gene names | GPR125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
HABP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.001970 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14520, O00663 | Gene names | HABP2, HGFAL, PHBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) (Hepatocyte growth factor activator-like protein) (Factor VII-activating protease) (Factor seven-activating protease) (FSAP) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.008079 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
NR1H4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.013934 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60641, Q60642, Q60643 | Gene names | Nr1h4, Bar, Fxr, Rip14 | |||
|
Domain Architecture |

|
|||||
| Description | Bile acid receptor (Farnesoid X-activated receptor) (Farnesol receptor HRR-1) (Retinoid X receptor-interacting protein 14) (RXR-interacting protein 14). | |||||
|
ARVC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.016102 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.034155 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.033917 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
C1QRF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.008528 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75973 | Gene names | C1QL1, C1QRF, CRF | |||
|
Domain Architecture |
|
|||||
| Description | C1q-related factor precursor (Complement component 1 Q subcomponent- like 1). | |||||
|
CJ120_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.034189 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SQS8 | Gene names | C10orf120 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf120. | |||||
|
K0460_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.021076 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
RING1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.008215 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.025819 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.014367 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.016922 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
T240L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.017000 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.006069 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
C1QRF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.007649 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88992 | Gene names | C1ql1, C1qrf, Crf | |||
|
Domain Architecture |
|
|||||
| Description | C1q-related factor precursor (Complement component 1 Q subcomponent- like 1). | |||||
|
CD44_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.016050 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15379, Q05732, Q61395, Q62060, Q62061, Q62062, Q62063, Q62408, Q62409, Q64296, Q99J14, Q9QYX8 | Gene names | Cd44, Ly-24 | |||
|
Domain Architecture |
|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein 1) (PGP-1) (HUTCH-I) (Extracellular matrix receptor III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Lymphocyte antigen Ly-24). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.018041 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
COT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.006434 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24468, Q03754 | Gene names | NR2F2, ARP1, TFCOUP2 | |||
|
Domain Architecture |
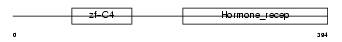
|
|||||
| Description | COUP transcription factor 2 (COUP-TF2) (COUP-TF II) (Apolipoprotein AI regulatory protein 1) (ARP-1). | |||||
|
COT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.006434 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43135 | Gene names | Nr2f2, Aporp1, Arp-1, Arp1, Tfcoup2 | |||
|
Domain Architecture |
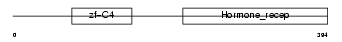
|
|||||
| Description | COUP transcription factor 2 (COUP-TF2) (COUP-TF II) (Apolipoprotein AI regulatory protein 1) (ARP-1). | |||||
|
DC1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.015975 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6G9, Q53HC8, Q53HK7 | Gene names | DYNC1LI1, DNCLI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
FOXD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.011909 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |
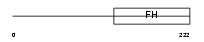
|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.032924 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.003980 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.010723 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
POGZ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.001263 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.008087 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SFR16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.027704 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.033987 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.024685 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
UBE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.007911 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
UN84A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.008514 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D666, Q80SU8, Q8BZ99, Q99P23 | Gene names | Unc84a, Sun1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sad1/unc-84 protein-like 1 (Unc-84 homolog A). | |||||
|
CAND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.058605 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
K0423_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.063154 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
K0423_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.067068 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
CLAP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CLAP2_HUMAN
|
||||||
| NC score | 0.991846 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O75122, Q7L8F6, Q8N6R6, Q9BQT3, Q9BQT4, Q9H7A3, Q9NSZ2 | Gene names | CLASP2, KIAA0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CLAP1_HUMAN
|
||||||
| NC score | 0.957978 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
CKAP5_HUMAN
|
||||||
| NC score | 0.179794 (rank : 4) | θ value | 0.0148317 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14008, Q14668 | Gene names | CKAP5, KIAA0097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 5 (Colonic and hepatic tumor over- expressed protein) (Ch-TOG protein). | |||||
|
CAND1_HUMAN
|
||||||
| NC score | 0.117960 (rank : 5) | θ value | 0.0148317 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86VP6, O94918, Q6PIY4, Q8NDJ4, Q96JZ9, Q96T19, Q9BTC4, Q9H0G2, Q9P0H7, Q9UF85 | Gene names | CAND1, KIAA0829, TIP120, TIP120A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1) (TBP-interacting protein TIP120A) (TBP-interacting protein of 120 kDa A). | |||||
|
IPO4_HUMAN
|
||||||
| NC score | 0.117744 (rank : 6) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TEX9, Q86TZ9, Q8NCG8, Q96SJ3, Q9BTI4, Q9H5L0 | Gene names | IPO4, IMP4B, RANBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-4 (Importin 4b) (Imp4b) (Ran-binding protein 4) (RanBP4). | |||||
|
CAND1_MOUSE
|
||||||
| NC score | 0.115976 (rank : 7) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZQ38, Q6PFR0, Q9CV45 | Gene names | Cand1, D10Ertd516e, Kiaa0829 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1). | |||||
|
ZRAB2_HUMAN
|
||||||
| NC score | 0.099008 (rank : 8) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
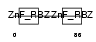
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
IPO4_MOUSE
|
||||||
| NC score | 0.096806 (rank : 9) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
CAND2_HUMAN
|
||||||
| NC score | 0.089139 (rank : 10) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75155 | Gene names | CAND2, KIAA0667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2). | |||||
|
TEX10_HUMAN
|
||||||
| NC score | 0.072296 (rank : 11) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXF1, Q5T722, Q5T723, Q8NCN8, Q8TDY0 | Gene names | TEX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 10 protein. | |||||
|
K0423_MOUSE
|
||||||
| NC score | 0.067068 (rank : 12) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.063154 (rank : 13) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
CAND2_MOUSE
|
||||||
| NC score | 0.058605 (rank : 14) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
UB2J1_MOUSE
|
||||||
| NC score | 0.052063 (rank : 15) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJZ4, Q9DC92 | Gene names | Ube2j1, Ncube1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 J1 (EC 6.3.2.19) (Non-canonical ubiquitin-conjugating enzyme 1) (NCUBE1). | |||||
|
FOXN3_MOUSE
|
||||||
| NC score | 0.048444 (rank : 16) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q499D0, Q8C5N8, Q9CU83 | Gene names | Ches1, Foxn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.048173 (rank : 17) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
FOXN3_HUMAN
|
||||||
| NC score | 0.045879 (rank : 18) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00409, Q96II7, Q9UIE7 | Gene names | CHES1, FOXN3 | |||
|
Domain Architecture |

|
|||||
| Description | Checkpoint suppressor 1 (Forkhead box protein N3). | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.045752 (rank : 19) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.043513 (rank : 20) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.042925 (rank : 21) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
TIRAP_HUMAN
|
||||||
| NC score | 0.042069 (rank : 22) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58753, Q8N5E5 | Gene names | TIRAP, MAL | |||
|
Domain Architecture |

|
|||||
| Description | Toll-interleukin 1 receptor domain-containing adapter protein (TIR domain-containing adapter protein) (MyD88 adapter-like protein) (Adaptor protein Wyatt). | |||||
|
PRC1_MOUSE
|
||||||
| NC score | 0.041832 (rank : 23) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
EPIPL_MOUSE
|
||||||
| NC score | 0.039257 (rank : 24) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0W0 | Gene names | Eppk1 | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin. | |||||
|
ANFC_HUMAN
|
||||||
| NC score | 0.037067 (rank : 25) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23582 | Gene names | NPPC, CNP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-type natriuretic peptide precursor [Contains: CNP-22; CNP-29; CNP- 53]. | |||||
|
MIA40_MOUSE
|
||||||
| NC score | 0.034217 (rank : 26) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEA4 | Gene names | Chchd4, Mia40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial intermembrane space import and assembly protein 40 (Coiled-coil-helix-coiled-coil-helix domain-containing protein 4). | |||||
|
CJ120_HUMAN
|
||||||
| NC score | 0.034189 (rank : 27) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SQS8 | Gene names | C10orf120 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf120. | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.034155 (rank : 28) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.033987 (rank : 29) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.033917 (rank : 30) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.032924 (rank : 31) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
ATG9A_HUMAN
|
||||||
| NC score | 0.030028 (rank : 32) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3C6, Q6P0N7, Q7Z317, Q7Z320, Q8NDK6, Q8WU65, Q9BVL5, Q9H6L1, Q9HAG7 | Gene names | ATG9A, APG9L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 9A (APG9-like 1). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.029610 (rank : 33) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
K1949_HUMAN
|
||||||
| NC score | 0.028023 (rank : 34) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NYC8, Q68CK8, Q6ZUJ6, Q8NDQ4, Q8TF52, Q9BRL9 | Gene names | KIAA1949 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949. | |||||
|
COVA1_MOUSE
|
||||||
| NC score | 0.027789 (rank : 35) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
SFR16_HUMAN
|
||||||
| NC score | 0.027704 (rank : 36) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
MAF_HUMAN
|
||||||
| NC score | 0.027429 (rank : 37) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.025819 (rank : 38) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
MAF_MOUSE
|
||||||
| NC score | 0.024685 (rank : 39) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.024685 (rank : 40) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.024160 (rank : 41) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.022773 (rank : 42) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CDR2_MOUSE
|
||||||
| NC score | 0.022555 (rank : 43) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97817 | Gene names | Cdr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2. | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.021076 (rank : 44) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
NR1H4_HUMAN
|
||||||
| NC score | 0.020337 (rank : 45) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RI1, Q8NFP5, Q8NFP6, Q92943 | Gene names | NR1H4, BAR, FXR, HRR1, RIP14 | |||
|
Domain Architecture |

|
|||||
| Description | Bile acid receptor (Farnesoid X-activated receptor) (Farnesol receptor HRR-1) (Retinoid X receptor-interacting protein 14) (RXR-interacting protein 14). | |||||
|
ZSWM5_HUMAN
|
||||||
| NC score | 0.019456 (rank : 46) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P217, Q5SXQ9 | Gene names | ZSWIM5, KIAA1511 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 5. | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.018995 (rank : 47) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.018223 (rank : 48) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.018041 (rank : 49) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
RBP26_HUMAN
|
||||||
| NC score | 0.017589 (rank : 50) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.017218 (rank : 51) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
T240L_MOUSE
|
||||||
| NC score | 0.017000 (rank : 52) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.016922 (rank : 53) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
TSH3_MOUSE
|
||||||
| NC score | 0.016589 (rank : 54) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.016102 (rank : 55) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
CD44_MOUSE
|
||||||
| NC score | 0.016050 (rank : 56) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15379, Q05732, Q61395, Q62060, Q62061, Q62062, Q62063, Q62408, Q62409, Q64296, Q99J14, Q9QYX8 | Gene names | Cd44, Ly-24 | |||
|
Domain Architecture |

|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein 1) (PGP-1) (HUTCH-I) (Extracellular matrix receptor III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Lymphocyte antigen Ly-24). | |||||
|
YYAP1_HUMAN
|
||||||
| NC score | 0.015983 (rank : 57) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H869, Q5VYZ4, Q5VYZ7, Q7L4C3, Q7L5E2, Q8IXA6, Q8TEW5, Q8TF04, Q96HB6, Q9BQ64, Q9NV84 | Gene names | YY1AP1, HCCA2, YY1AP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YY1-associated protein 1 (Hepatocellular carcinoma susceptibility protein) (Hepatocellular carcinoma-associated protein 2). | |||||
|
DC1L1_HUMAN
|
||||||
| NC score | 0.015975 (rank : 58) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6G9, Q53HC8, Q53HK7 | Gene names | DYNC1LI1, DNCLI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
CASR_MOUSE
|
||||||
| NC score | 0.015063 (rank : 59) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY96, O08968, O88519, Q9QY95, Q9QZU8, Q9R1D6, Q9R1Y2 | Gene names | Casr, Gprc2a | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.014780 (rank : 60) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.014431 (rank : 61) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.014367 (rank : 62) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
NR1H4_MOUSE
|
||||||
| NC score | 0.013934 (rank : 63) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60641, Q60642, Q60643 | Gene names | Nr1h4, Bar, Fxr, Rip14 | |||
|
Domain Architecture |

|
|||||
| Description | Bile acid receptor (Farnesoid X-activated receptor) (Farnesol receptor HRR-1) (Retinoid X receptor-interacting protein 14) (RXR-interacting protein 14). | |||||
|
GP125_HUMAN
|
||||||
| NC score | 0.013261 (rank : 64) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWK6, Q6UXK9, Q86SQ5, Q8TC55 | Gene names | GPR125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
GCP60_HUMAN
|
||||||
| NC score | 0.013093 (rank : 65) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.012839 (rank : 66) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
OST6_HUMAN
|
||||||
| NC score | 0.012276 (rank : 67) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96QI5, Q96RX7 | Gene names | HS3ST6, HS3ST5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan sulfate glucosamine 3-O-sulfotransferase 6 (EC 2.8.2.23) (Heparan sulfate D-glucosaminyl 3-O-sulfotransferase 6) (Heparan sulfate 3-O-sulfotransferase 6) (h3-OST-6). | |||||
|
FOXD1_MOUSE
|
||||||
| NC score | 0.011909 (rank : 68) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
TLN2_MOUSE
|
||||||
| NC score | 0.011279 (rank : 69) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71LX4, Q8BWK0 | Gene names | Tln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Talin-2. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.011252 (rank : 70) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.010723 (rank : 71) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
BTBD2_HUMAN
|
||||||
| NC score | 0.009740 (rank : 72) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX70, O60418, O75248, Q7Z5W0, Q96SX8, Q9NPS1, Q9NX81 | Gene names | BTBD2 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 2. | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.009701 (rank : 73) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.009672 (rank : 74) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
C1QRF_HUMAN
|
||||||
| NC score | 0.008528 (rank : 75) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75973 | Gene names | C1QL1, C1QRF, CRF | |||
|
Domain Architecture |

|
|||||
| Description | C1q-related factor precursor (Complement component 1 Q subcomponent- like 1). | |||||
|
UN84A_MOUSE
|
||||||
| NC score | 0.008514 (rank : 76) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D666, Q80SU8, Q8BZ99, Q99P23 | Gene names | Unc84a, Sun1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sad1/unc-84 protein-like 1 (Unc-84 homolog A). | |||||
|
RING1_MOUSE
|
||||||
| NC score | 0.008215 (rank : 77) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.008087 (rank : 78) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.008079 (rank : 79) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
UBE1_MOUSE
|
||||||
| NC score | 0.007911 (rank : 80) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02053, Q542H8 | Gene names | Ube1x, Uba1, Ube1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-activating enzyme E1 1. | |||||
|
C1QRF_MOUSE
|
||||||
| NC score | 0.007649 (rank : 81) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88992 | Gene names | C1ql1, C1qrf, Crf | |||
|
Domain Architecture |

|
|||||
| Description | C1q-related factor precursor (Complement component 1 Q subcomponent- like 1). | |||||
|
COT2_HUMAN
|
||||||
| NC score | 0.006434 (rank : 82) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24468, Q03754 | Gene names | NR2F2, ARP1, TFCOUP2 | |||
|
Domain Architecture |

|
|||||
| Description | COUP transcription factor 2 (COUP-TF2) (COUP-TF II) (Apolipoprotein AI regulatory protein 1) (ARP-1). | |||||
|
COT2_MOUSE
|
||||||
| NC score | 0.006434 (rank : 83) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43135 | Gene names | Nr2f2, Aporp1, Arp-1, Arp1, Tfcoup2 | |||
|
Domain Architecture |

|
|||||
| Description | COUP transcription factor 2 (COUP-TF2) (COUP-TF II) (Apolipoprotein AI regulatory protein 1) (ARP-1). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.006069 (rank : 84) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CD2L6_MOUSE
|
||||||
| NC score | 0.004086 (rank : 85) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BWD8, Q80TM1 | Gene names | Cdc2l6, Kiaa1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6). | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.003980 (rank : 86) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
SP8_MOUSE
|
||||||
| NC score | 0.003602 (rank : 87) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMJ8, Q8BWA3 | Gene names | Sp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp8. | |||||
|
MARK3_HUMAN
|
||||||
| NC score | 0.002567 (rank : 88) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
HABP2_HUMAN
|
||||||
| NC score | 0.001970 (rank : 89) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14520, O00663 | Gene names | HABP2, HGFAL, PHBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) (Hepatocyte growth factor activator-like protein) (Factor VII-activating protease) (Factor seven-activating protease) (FSAP) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
FIZ1_MOUSE
|
||||||
| NC score | 0.001750 (rank : 90) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTJ4, Q91W14, Q9CTG3 | Gene names | Fiz1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flt3-interacting zinc finger protein 1. | |||||
|
POGZ_MOUSE
|
||||||
| NC score | 0.001263 (rank : 91) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||