Please be patient as the page loads
|
UBP53_HUMAN
|
||||||
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBP53_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.944994 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
UBP54_MOUSE
|
||||||
| θ value | 3.39372e-140 (rank : 3) | NC score | 0.953313 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP54_HUMAN
|
||||||
| θ value | 4.43234e-140 (rank : 4) | NC score | 0.953092 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.183495 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.125463 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.102556 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.098739 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.098806 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.046775 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.017530 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.090968 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.093047 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.109268 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.021954 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.034889 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.091330 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.011431 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.065735 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.013351 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
M3K4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.004304 (rank : 76) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
LV1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.012129 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01724 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda-1 chain V regions MOPC 104E/RPC20/J558/S104 precursor. | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.017529 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
ITA5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.014354 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08648, Q96HA5 | Gene names | ITGA5, FNRA | |||
|
Domain Architecture |
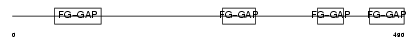
|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
JARD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.031830 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.053404 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.027571 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.011686 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.022747 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.011626 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.052682 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.079355 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
ZMYM6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.020763 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.045273 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
BCL7A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.022249 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
DC1I1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.013768 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14576, Q9Y2X1 | Gene names | DYNC1I1, DNCI1, DNCIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 1 (Dynein intermediate chain 1, cytosolic) (DH IC-1) (Cytoplasmic dynein intermediate chain 1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.055331 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LV1E_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.010937 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01727 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda-1 chain V region S43 precursor. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.028854 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ARX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.004298 (rank : 77) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.023899 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
LRCH3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.007198 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU0, Q3U222, Q3UZ74 | Gene names | Lrch3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.006140 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SPAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.028899 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UM82, O94857 | Gene names | SPATA2, KIAA0757, PD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 2 (Spermatogenesis-associated protein PD1). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.056075 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.056064 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
ZN599_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | -0.000965 (rank : 80) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NL3, Q569K0, Q5PRG1 | Gene names | ZNF599 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 599. | |||||
|
BNI3L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.023258 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60238 | Gene names | BNIP3L, BNIP3A, BNIP3H, NIX | |||
|
Domain Architecture |
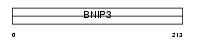
|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like (NIP3L) (NIP3-like protein X) (BCL2/adenovirus E1B 19 kDa protein-interacting protein 3A). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.008636 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.008062 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
ITA5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.012113 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11688 | Gene names | Itga5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
KNTC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.015824 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50748 | Gene names | KNTC1, KIAA0166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore-associated protein 1 (Rough deal homolog) (hRod) (HsROD) (Rod). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.054359 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.021536 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BNI3L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.021392 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2F7, Q545J6 | Gene names | Bnip3l, Nix | |||
|
Domain Architecture |
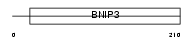
|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like (NIP3L) (NIP3-like protein X). | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.007283 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.010261 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
LV1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.010683 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01723 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda-1 chain V region precursor. | |||||
|
PKN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.001098 (rank : 79) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5Z2, Q9UM03 | Gene names | PKN3, PKNBETA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.012031 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
TNF13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.015768 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75888, Q96HV6, Q9P1M8, Q9P1M9 | Gene names | TNFSF13, APRIL, TALL2, ZTNF2 | |||
|
Domain Architecture |
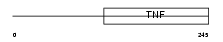
|
|||||
| Description | Tumor necrosis factor ligand superfamily member 13 precursor (A proliferation-inducing ligand) (APRIL) (TNF- and APOL-related leukocyte expressed ligand 2) (TALL-2) (TNF-related death ligand 1) (TRDL-1) (CD256 antigen). | |||||
|
DZIP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.008706 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.010097 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
LV1D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.008780 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01726 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda-1 chain V region H2020 precursor. | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.010506 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.010532 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NELL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.001882 (rank : 78) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.014394 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
TADBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.011147 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13148, Q96DJ0 | Gene names | TARDBP, TDP43 | |||
|
Domain Architecture |
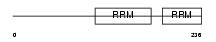
|
|||||
| Description | TAR DNA-binding protein 43 (TDP-43). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.012246 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.056040 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UB17L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.050436 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.055024 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.057148 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.062318 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.062318 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.051101 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052009 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.065548 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.051577 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP53_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
UBP54_MOUSE
|
||||||
| NC score | 0.953313 (rank : 2) | θ value | 3.39372e-140 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP54_HUMAN
|
||||||
| NC score | 0.953092 (rank : 3) | θ value | 4.43234e-140 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.944994 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.183495 (rank : 5) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.125463 (rank : 6) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.109268 (rank : 7) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.102556 (rank : 8) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.098806 (rank : 9) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.098739 (rank : 10) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.093047 (rank : 11) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.091330 (rank : 12) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.090968 (rank : 13) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.079355 (rank : 14) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.065735 (rank : 15) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.065548 (rank : 16) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.062318 (rank : 17) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.062318 (rank : 18) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.057148 (rank : 19) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.056075 (rank : 20) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.056064 (rank : 21) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.056040 (rank : 22) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.055331 (rank : 23) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.055024 (rank : 24) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.054359 (rank : 25) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.053404 (rank : 26) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.052682 (rank : 27) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.052009 (rank : 28) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.051577 (rank : 29) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.051101 (rank : 30) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.050436 (rank : 31) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.046775 (rank : 32) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.045273 (rank : 33) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.034889 (rank : 34) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.031830 (rank : 35) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
SPAT2_HUMAN
|
||||||
| NC score | 0.028899 (rank : 36) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UM82, O94857 | Gene names | SPATA2, KIAA0757, PD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 2 (Spermatogenesis-associated protein PD1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.028854 (rank : 37) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.027571 (rank : 38) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.023899 (rank : 39) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
BNI3L_HUMAN
|
||||||
| NC score | 0.023258 (rank : 40) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60238 | Gene names | BNIP3L, BNIP3A, BNIP3H, NIX | |||
|
Domain Architecture |

|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like (NIP3L) (NIP3-like protein X) (BCL2/adenovirus E1B 19 kDa protein-interacting protein 3A). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.022747 (rank : 41) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
BCL7A_HUMAN
|
||||||
| NC score | 0.022249 (rank : 42) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.021954 (rank : 43) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.021536 (rank : 44) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BNI3L_MOUSE
|
||||||
| NC score | 0.021392 (rank : 45) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2F7, Q545J6 | Gene names | Bnip3l, Nix | |||
|
Domain Architecture |

|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like (NIP3L) (NIP3-like protein X). | |||||
|
ZMYM6_HUMAN
|
||||||
| NC score | 0.020763 (rank : 46) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95789, Q96IY0 | Gene names | ZMYM6, ZNF258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 6 (Zinc finger protein 258). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.017530 (rank : 47) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.017529 (rank : 48) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
KNTC1_HUMAN
|
||||||
| NC score | 0.015824 (rank : 49) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50748 | Gene names | KNTC1, KIAA0166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore-associated protein 1 (Rough deal homolog) (hRod) (HsROD) (Rod). | |||||
|
TNF13_HUMAN
|
||||||
| NC score | 0.015768 (rank : 50) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75888, Q96HV6, Q9P1M8, Q9P1M9 | Gene names | TNFSF13, APRIL, TALL2, ZTNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor ligand superfamily member 13 precursor (A proliferation-inducing ligand) (APRIL) (TNF- and APOL-related leukocyte expressed ligand 2) (TALL-2) (TNF-related death ligand 1) (TRDL-1) (CD256 antigen). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.014394 (rank : 51) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
ITA5_HUMAN
|
||||||
| NC score | 0.014354 (rank : 52) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08648, Q96HA5 | Gene names | ITGA5, FNRA | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
DC1I1_HUMAN
|
||||||
| NC score | 0.013768 (rank : 53) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14576, Q9Y2X1 | Gene names | DYNC1I1, DNCI1, DNCIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 1 (Dynein intermediate chain 1, cytosolic) (DH IC-1) (Cytoplasmic dynein intermediate chain 1). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.013351 (rank : 54) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.012246 (rank : 55) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
LV1B_MOUSE
|
||||||
| NC score | 0.012129 (rank : 56) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01724 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda-1 chain V regions MOPC 104E/RPC20/J558/S104 precursor. | |||||
|
ITA5_MOUSE
|
||||||
| NC score | 0.012113 (rank : 57) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11688 | Gene names | Itga5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.012031 (rank : 58) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.011686 (rank : 59) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.011626 (rank : 60) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | 0.011431 (rank : 61) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
TADBP_HUMAN
|
||||||
| NC score | 0.011147 (rank : 62) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13148, Q96DJ0 | Gene names | TARDBP, TDP43 | |||
|
Domain Architecture |

|
|||||
| Description | TAR DNA-binding protein 43 (TDP-43). | |||||
|
LV1E_MOUSE
|
||||||
| NC score | 0.010937 (rank : 63) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01727 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda-1 chain V region S43 precursor. | |||||
|
LV1A_MOUSE
|
||||||
| NC score | 0.010683 (rank : 64) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01723 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda-1 chain V region precursor. | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.010532 (rank : 65) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.010506 (rank : 66) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.010261 (rank : 67) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.010097 (rank : 68) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
LV1D_MOUSE
|
||||||
| NC score | 0.008780 (rank : 69) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01726 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda-1 chain V region H2020 precursor. | |||||
|
DZIP3_MOUSE
|
||||||
| NC score | 0.008706 (rank : 70) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.008636 (rank : 71) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.008062 (rank : 72) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.007283 (rank : 73) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
LRCH3_MOUSE
|
||||||
| NC score | 0.007198 (rank : 74) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU0, Q3U222, Q3UZ74 | Gene names | Lrch3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.006140 (rank : 75) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
M3K4_MOUSE
|
||||||
| NC score | 0.004304 (rank : 76) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
ARX_HUMAN
|
||||||
| NC score | 0.004298 (rank : 77) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
NELL2_MOUSE
|
||||||
| NC score | 0.001882 (rank : 78) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
PKN3_HUMAN
|
||||||
| NC score | 0.001098 (rank : 79) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5Z2, Q9UM03 | Gene names | PKN3, PKNBETA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
ZN599_HUMAN
|
||||||
| NC score | -0.000965 (rank : 80) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96NL3, Q569K0, Q5PRG1 | Gene names | ZNF599 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 599. | |||||